
Aliiroseovarius crassostreae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Aliiroseovarius
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
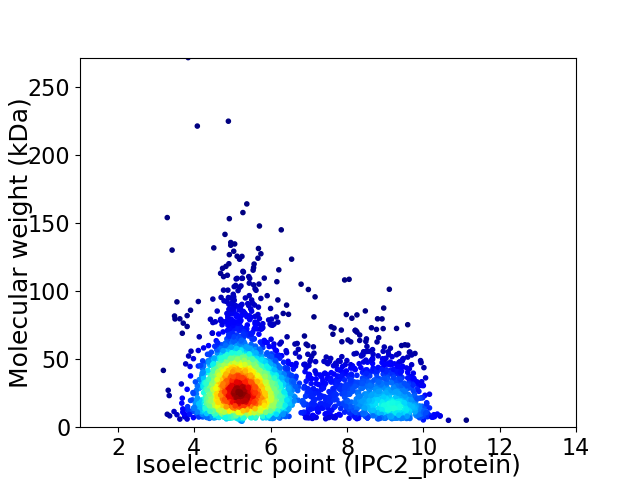
Virtual 2D-PAGE plot for 3465 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
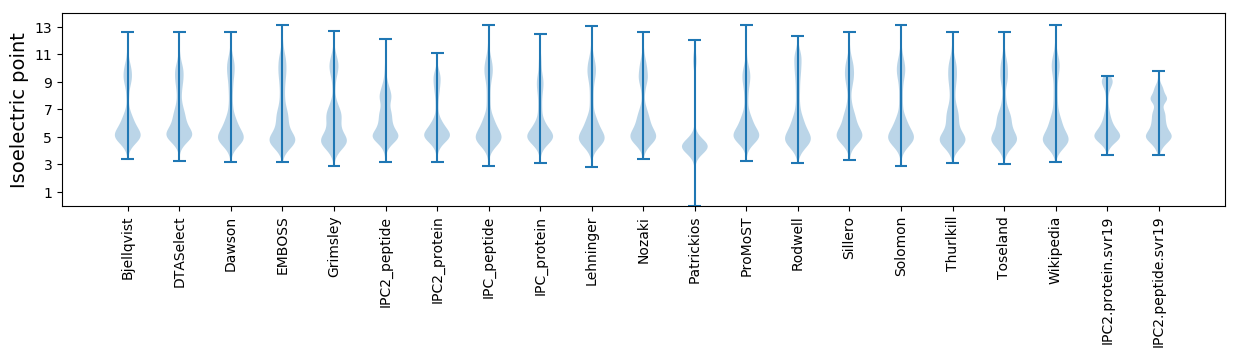
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P7ITD1|A0A0P7ITD1_9RHOB L-aspartate oxidase OS=Aliiroseovarius crassostreae OX=154981 GN=AKJ29_07290 PE=3 SV=1
MM1 pKa = 6.98ATGYY5 pKa = 10.22LVQLGDD11 pKa = 3.71SSLDD15 pKa = 3.4SGDD18 pKa = 4.96AIIGPTTSFTIDD30 pKa = 3.48TILGSGSWTWSGTYY44 pKa = 10.58GGTTYY49 pKa = 10.83TNEE52 pKa = 4.38TEE54 pKa = 3.85TGTYY58 pKa = 10.6YY59 pKa = 10.88LGSDD63 pKa = 3.34GNIYY67 pKa = 8.37FTSNNGPVDD76 pKa = 4.39TISSASVISAPSYY89 pKa = 9.53STADD93 pKa = 3.55DD94 pKa = 3.87VLTGGDD100 pKa = 3.69GADD103 pKa = 3.91VIDD106 pKa = 4.36TSFTDD111 pKa = 3.53EE112 pKa = 5.28DD113 pKa = 4.36GEE115 pKa = 4.65KK116 pKa = 10.58VDD118 pKa = 4.95GGDD121 pKa = 4.22GATGDD126 pKa = 3.73NADD129 pKa = 4.08VVDD132 pKa = 5.39AGAGDD137 pKa = 4.64DD138 pKa = 4.02YY139 pKa = 12.1VEE141 pKa = 4.83AGYY144 pKa = 11.36GDD146 pKa = 3.6DD147 pKa = 4.21TIYY150 pKa = 11.2GGTGSDD156 pKa = 3.77TIEE159 pKa = 4.65GGDD162 pKa = 4.18GNDD165 pKa = 4.69SIYY168 pKa = 11.18GDD170 pKa = 4.89DD171 pKa = 4.8DD172 pKa = 4.67LSSVTPEE179 pKa = 4.95PITIHH184 pKa = 6.3EE185 pKa = 4.83GNFTDD190 pKa = 3.59TTNGYY195 pKa = 7.54TVTAQNVVGGSLTTASSSNIATYY218 pKa = 10.71SGGGFGASGTVSDD231 pKa = 4.75SDD233 pKa = 4.2SSVTSQIGYY242 pKa = 10.02DD243 pKa = 3.59LASGLSEE250 pKa = 4.08NVTVTLDD257 pKa = 3.76GEE259 pKa = 4.09ASEE262 pKa = 5.45ASFSFANLYY271 pKa = 9.2TSTYY275 pKa = 10.98AEE277 pKa = 4.23AGHH280 pKa = 5.78WAIYY284 pKa = 10.21NDD286 pKa = 3.54GVLVAEE292 pKa = 5.26GDD294 pKa = 3.82FTEE297 pKa = 4.57QGAGTGSGTVDD308 pKa = 2.89ISGHH312 pKa = 5.38GNFDD316 pKa = 3.7QIVFTALPQTDD327 pKa = 3.7GTDD330 pKa = 3.25GSDD333 pKa = 3.42YY334 pKa = 11.01HH335 pKa = 6.05LTSVSFTPVPVDD347 pKa = 4.39PIAGNDD353 pKa = 3.78SLSGGAGDD361 pKa = 6.18DD362 pKa = 4.4FIDD365 pKa = 5.03GNGGDD370 pKa = 4.03DD371 pKa = 3.75TLTGGSGSDD380 pKa = 3.43TLHH383 pKa = 6.7GGDD386 pKa = 4.58GEE388 pKa = 4.58DD389 pKa = 4.87LIHH392 pKa = 7.06VGAGDD397 pKa = 3.72TASGGTGSDD406 pKa = 2.88RR407 pKa = 11.84FEE409 pKa = 5.45LNPTDD414 pKa = 4.71ALDD417 pKa = 4.13GSGPTITLDD426 pKa = 3.38GGEE429 pKa = 4.91DD430 pKa = 3.95ADD432 pKa = 6.08DD433 pKa = 4.44SDD435 pKa = 6.11IDD437 pKa = 3.93TLDD440 pKa = 3.81LKK442 pKa = 11.41GLVNDD447 pKa = 4.26WSNVVFDD454 pKa = 5.27PDD456 pKa = 3.46NPEE459 pKa = 4.07NGTATLSDD467 pKa = 3.59GTVVTFTNFEE477 pKa = 4.03QIIICFTGSTRR488 pKa = 11.84IEE490 pKa = 4.4TPFGPRR496 pKa = 11.84PVQDD500 pKa = 4.46LRR502 pKa = 11.84PGDD505 pKa = 4.59LIITRR510 pKa = 11.84DD511 pKa = 3.51NGLQPLLWIGKK522 pKa = 7.44SHH524 pKa = 6.65VAAQGRR530 pKa = 11.84LAPIRR535 pKa = 11.84FCKK538 pKa = 9.99GAIGNSRR545 pKa = 11.84PLLVSPQHH553 pKa = 6.77RR554 pKa = 11.84ILLKK558 pKa = 10.54SPKK561 pKa = 9.16ATLLFDD567 pKa = 3.74TPEE570 pKa = 4.04VLVPACHH577 pKa = 6.52MINGEE582 pKa = 4.21TIYY585 pKa = 10.78RR586 pKa = 11.84EE587 pKa = 3.97AASEE591 pKa = 3.93VTYY594 pKa = 10.79YY595 pKa = 10.98HH596 pKa = 7.17LLFDD600 pKa = 3.63QHH602 pKa = 5.85EE603 pKa = 4.68VIYY606 pKa = 10.88SEE608 pKa = 4.42GVPTEE613 pKa = 4.29SFHH616 pKa = 6.8PAWPTLGGLDD626 pKa = 3.36TQTRR630 pKa = 11.84EE631 pKa = 3.74EE632 pKa = 4.18VLALFPALRR641 pKa = 11.84CSNTGYY647 pKa = 11.05GKK649 pKa = 7.71TARR652 pKa = 11.84SVLRR656 pKa = 11.84RR657 pKa = 11.84YY658 pKa = 7.42EE659 pKa = 4.05TRR661 pKa = 11.84LLLAGG666 pKa = 4.15
MM1 pKa = 6.98ATGYY5 pKa = 10.22LVQLGDD11 pKa = 3.71SSLDD15 pKa = 3.4SGDD18 pKa = 4.96AIIGPTTSFTIDD30 pKa = 3.48TILGSGSWTWSGTYY44 pKa = 10.58GGTTYY49 pKa = 10.83TNEE52 pKa = 4.38TEE54 pKa = 3.85TGTYY58 pKa = 10.6YY59 pKa = 10.88LGSDD63 pKa = 3.34GNIYY67 pKa = 8.37FTSNNGPVDD76 pKa = 4.39TISSASVISAPSYY89 pKa = 9.53STADD93 pKa = 3.55DD94 pKa = 3.87VLTGGDD100 pKa = 3.69GADD103 pKa = 3.91VIDD106 pKa = 4.36TSFTDD111 pKa = 3.53EE112 pKa = 5.28DD113 pKa = 4.36GEE115 pKa = 4.65KK116 pKa = 10.58VDD118 pKa = 4.95GGDD121 pKa = 4.22GATGDD126 pKa = 3.73NADD129 pKa = 4.08VVDD132 pKa = 5.39AGAGDD137 pKa = 4.64DD138 pKa = 4.02YY139 pKa = 12.1VEE141 pKa = 4.83AGYY144 pKa = 11.36GDD146 pKa = 3.6DD147 pKa = 4.21TIYY150 pKa = 11.2GGTGSDD156 pKa = 3.77TIEE159 pKa = 4.65GGDD162 pKa = 4.18GNDD165 pKa = 4.69SIYY168 pKa = 11.18GDD170 pKa = 4.89DD171 pKa = 4.8DD172 pKa = 4.67LSSVTPEE179 pKa = 4.95PITIHH184 pKa = 6.3EE185 pKa = 4.83GNFTDD190 pKa = 3.59TTNGYY195 pKa = 7.54TVTAQNVVGGSLTTASSSNIATYY218 pKa = 10.71SGGGFGASGTVSDD231 pKa = 4.75SDD233 pKa = 4.2SSVTSQIGYY242 pKa = 10.02DD243 pKa = 3.59LASGLSEE250 pKa = 4.08NVTVTLDD257 pKa = 3.76GEE259 pKa = 4.09ASEE262 pKa = 5.45ASFSFANLYY271 pKa = 9.2TSTYY275 pKa = 10.98AEE277 pKa = 4.23AGHH280 pKa = 5.78WAIYY284 pKa = 10.21NDD286 pKa = 3.54GVLVAEE292 pKa = 5.26GDD294 pKa = 3.82FTEE297 pKa = 4.57QGAGTGSGTVDD308 pKa = 2.89ISGHH312 pKa = 5.38GNFDD316 pKa = 3.7QIVFTALPQTDD327 pKa = 3.7GTDD330 pKa = 3.25GSDD333 pKa = 3.42YY334 pKa = 11.01HH335 pKa = 6.05LTSVSFTPVPVDD347 pKa = 4.39PIAGNDD353 pKa = 3.78SLSGGAGDD361 pKa = 6.18DD362 pKa = 4.4FIDD365 pKa = 5.03GNGGDD370 pKa = 4.03DD371 pKa = 3.75TLTGGSGSDD380 pKa = 3.43TLHH383 pKa = 6.7GGDD386 pKa = 4.58GEE388 pKa = 4.58DD389 pKa = 4.87LIHH392 pKa = 7.06VGAGDD397 pKa = 3.72TASGGTGSDD406 pKa = 2.88RR407 pKa = 11.84FEE409 pKa = 5.45LNPTDD414 pKa = 4.71ALDD417 pKa = 4.13GSGPTITLDD426 pKa = 3.38GGEE429 pKa = 4.91DD430 pKa = 3.95ADD432 pKa = 6.08DD433 pKa = 4.44SDD435 pKa = 6.11IDD437 pKa = 3.93TLDD440 pKa = 3.81LKK442 pKa = 11.41GLVNDD447 pKa = 4.26WSNVVFDD454 pKa = 5.27PDD456 pKa = 3.46NPEE459 pKa = 4.07NGTATLSDD467 pKa = 3.59GTVVTFTNFEE477 pKa = 4.03QIIICFTGSTRR488 pKa = 11.84IEE490 pKa = 4.4TPFGPRR496 pKa = 11.84PVQDD500 pKa = 4.46LRR502 pKa = 11.84PGDD505 pKa = 4.59LIITRR510 pKa = 11.84DD511 pKa = 3.51NGLQPLLWIGKK522 pKa = 7.44SHH524 pKa = 6.65VAAQGRR530 pKa = 11.84LAPIRR535 pKa = 11.84FCKK538 pKa = 9.99GAIGNSRR545 pKa = 11.84PLLVSPQHH553 pKa = 6.77RR554 pKa = 11.84ILLKK558 pKa = 10.54SPKK561 pKa = 9.16ATLLFDD567 pKa = 3.74TPEE570 pKa = 4.04VLVPACHH577 pKa = 6.52MINGEE582 pKa = 4.21TIYY585 pKa = 10.78RR586 pKa = 11.84EE587 pKa = 3.97AASEE591 pKa = 3.93VTYY594 pKa = 10.79YY595 pKa = 10.98HH596 pKa = 7.17LLFDD600 pKa = 3.63QHH602 pKa = 5.85EE603 pKa = 4.68VIYY606 pKa = 10.88SEE608 pKa = 4.42GVPTEE613 pKa = 4.29SFHH616 pKa = 6.8PAWPTLGGLDD626 pKa = 3.36TQTRR630 pKa = 11.84EE631 pKa = 3.74EE632 pKa = 4.18VLALFPALRR641 pKa = 11.84CSNTGYY647 pKa = 11.05GKK649 pKa = 7.71TARR652 pKa = 11.84SVLRR656 pKa = 11.84RR657 pKa = 11.84YY658 pKa = 7.42EE659 pKa = 4.05TRR661 pKa = 11.84LLLAGG666 pKa = 4.15
Molecular weight: 68.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P7JRR9|A0A0P7JRR9_9RHOB MPT synthase subunit 2 OS=Aliiroseovarius crassostreae OX=154981 GN=AKJ29_15895 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1035234 |
38 |
2637 |
298.8 |
32.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.435 ± 0.053 | 0.894 ± 0.014 |
6.069 ± 0.036 | 6.174 ± 0.037 |
3.815 ± 0.025 | 8.552 ± 0.044 |
2.109 ± 0.022 | 5.297 ± 0.028 |
3.778 ± 0.035 | 9.959 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.849 ± 0.021 | 2.856 ± 0.03 |
4.884 ± 0.031 | 3.393 ± 0.024 |
6.423 ± 0.041 | 5.36 ± 0.031 |
5.383 ± 0.028 | 7.142 ± 0.035 |
1.359 ± 0.017 | 2.267 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
