
Cyanoramphus nest associated circular K DNA virus
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.9
Get precalculated fractions of proteins
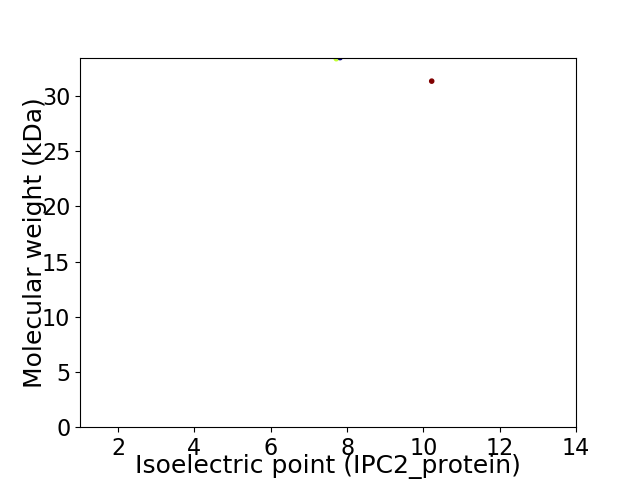
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L7UZA8|L7UZA8_9VIRU Replication associated protein OS=Cyanoramphus nest associated circular K DNA virus OX=1282444 PE=4 SV=1
MM1 pKa = 6.62NHH3 pKa = 5.27ARR5 pKa = 11.84ARR7 pKa = 11.84SKK9 pKa = 10.14NWCFTSYY16 pKa = 11.25DD17 pKa = 3.51EE18 pKa = 5.71SEE20 pKa = 4.41PDD22 pKa = 3.26LARR25 pKa = 11.84HH26 pKa = 5.19QEE28 pKa = 3.68HH29 pKa = 6.04LTYY32 pKa = 10.63YY33 pKa = 9.92IYY35 pKa = 10.65QPEE38 pKa = 4.15RR39 pKa = 11.84GVQGRR44 pKa = 11.84RR45 pKa = 11.84HH46 pKa = 4.49FQGYY50 pKa = 9.48AEE52 pKa = 4.06SRR54 pKa = 11.84EE55 pKa = 4.14RR56 pKa = 11.84IGVRR60 pKa = 11.84EE61 pKa = 4.19LQSFIGSRR69 pKa = 11.84AHH71 pKa = 6.96CEE73 pKa = 3.46IAFDD77 pKa = 4.25PEE79 pKa = 3.86ASRR82 pKa = 11.84LYY84 pKa = 10.2CRR86 pKa = 11.84KK87 pKa = 9.57EE88 pKa = 3.77STRR91 pKa = 11.84DD92 pKa = 3.29GVTTEE97 pKa = 3.41WGLFRR102 pKa = 11.84GKK104 pKa = 10.14KK105 pKa = 8.95QGSRR109 pKa = 11.84TDD111 pKa = 3.06IGTACDD117 pKa = 3.58TIRR120 pKa = 11.84RR121 pKa = 11.84SGLGACIKK129 pKa = 8.53EE130 pKa = 4.16HH131 pKa = 7.15PEE133 pKa = 3.97TFVKK137 pKa = 10.42YY138 pKa = 10.26HH139 pKa = 6.62GGLTIYY145 pKa = 10.3AAQQFDD151 pKa = 3.88PRR153 pKa = 11.84RR154 pKa = 11.84CTDD157 pKa = 3.57DD158 pKa = 4.06PPDD161 pKa = 3.21VRR163 pKa = 11.84IYY165 pKa = 11.13YY166 pKa = 10.26GVTGIGKK173 pKa = 7.62TRR175 pKa = 11.84SVYY178 pKa = 10.22AQYY181 pKa = 11.67GNDD184 pKa = 3.91IYY186 pKa = 11.37TKK188 pKa = 10.73DD189 pKa = 4.52DD190 pKa = 3.88SKK192 pKa = 10.71WWNGYY197 pKa = 8.63AAQKK201 pKa = 10.6CILFDD206 pKa = 3.63DD207 pKa = 4.17WVGSEE212 pKa = 4.39EE213 pKa = 3.99ISPVSLLKK221 pKa = 10.51ICDD224 pKa = 3.75RR225 pKa = 11.84YY226 pKa = 9.96PLQVQTKK233 pKa = 8.74GGYY236 pKa = 9.12VPLARR241 pKa = 11.84TTTVIIFTTTQHH253 pKa = 5.41EE254 pKa = 4.41NFWYY258 pKa = 10.45GGTKK262 pKa = 10.04HH263 pKa = 5.51EE264 pKa = 5.57ANWCSQRR271 pKa = 11.84AAWDD275 pKa = 3.41RR276 pKa = 11.84RR277 pKa = 11.84VSEE280 pKa = 5.42RR281 pKa = 11.84RR282 pKa = 11.84TEE284 pKa = 3.83WTQPP288 pKa = 3.13
MM1 pKa = 6.62NHH3 pKa = 5.27ARR5 pKa = 11.84ARR7 pKa = 11.84SKK9 pKa = 10.14NWCFTSYY16 pKa = 11.25DD17 pKa = 3.51EE18 pKa = 5.71SEE20 pKa = 4.41PDD22 pKa = 3.26LARR25 pKa = 11.84HH26 pKa = 5.19QEE28 pKa = 3.68HH29 pKa = 6.04LTYY32 pKa = 10.63YY33 pKa = 9.92IYY35 pKa = 10.65QPEE38 pKa = 4.15RR39 pKa = 11.84GVQGRR44 pKa = 11.84RR45 pKa = 11.84HH46 pKa = 4.49FQGYY50 pKa = 9.48AEE52 pKa = 4.06SRR54 pKa = 11.84EE55 pKa = 4.14RR56 pKa = 11.84IGVRR60 pKa = 11.84EE61 pKa = 4.19LQSFIGSRR69 pKa = 11.84AHH71 pKa = 6.96CEE73 pKa = 3.46IAFDD77 pKa = 4.25PEE79 pKa = 3.86ASRR82 pKa = 11.84LYY84 pKa = 10.2CRR86 pKa = 11.84KK87 pKa = 9.57EE88 pKa = 3.77STRR91 pKa = 11.84DD92 pKa = 3.29GVTTEE97 pKa = 3.41WGLFRR102 pKa = 11.84GKK104 pKa = 10.14KK105 pKa = 8.95QGSRR109 pKa = 11.84TDD111 pKa = 3.06IGTACDD117 pKa = 3.58TIRR120 pKa = 11.84RR121 pKa = 11.84SGLGACIKK129 pKa = 8.53EE130 pKa = 4.16HH131 pKa = 7.15PEE133 pKa = 3.97TFVKK137 pKa = 10.42YY138 pKa = 10.26HH139 pKa = 6.62GGLTIYY145 pKa = 10.3AAQQFDD151 pKa = 3.88PRR153 pKa = 11.84RR154 pKa = 11.84CTDD157 pKa = 3.57DD158 pKa = 4.06PPDD161 pKa = 3.21VRR163 pKa = 11.84IYY165 pKa = 11.13YY166 pKa = 10.26GVTGIGKK173 pKa = 7.62TRR175 pKa = 11.84SVYY178 pKa = 10.22AQYY181 pKa = 11.67GNDD184 pKa = 3.91IYY186 pKa = 11.37TKK188 pKa = 10.73DD189 pKa = 4.52DD190 pKa = 3.88SKK192 pKa = 10.71WWNGYY197 pKa = 8.63AAQKK201 pKa = 10.6CILFDD206 pKa = 3.63DD207 pKa = 4.17WVGSEE212 pKa = 4.39EE213 pKa = 3.99ISPVSLLKK221 pKa = 10.51ICDD224 pKa = 3.75RR225 pKa = 11.84YY226 pKa = 9.96PLQVQTKK233 pKa = 8.74GGYY236 pKa = 9.12VPLARR241 pKa = 11.84TTTVIIFTTTQHH253 pKa = 5.41EE254 pKa = 4.41NFWYY258 pKa = 10.45GGTKK262 pKa = 10.04HH263 pKa = 5.51EE264 pKa = 5.57ANWCSQRR271 pKa = 11.84AAWDD275 pKa = 3.41RR276 pKa = 11.84RR277 pKa = 11.84VSEE280 pKa = 5.42RR281 pKa = 11.84RR282 pKa = 11.84TEE284 pKa = 3.83WTQPP288 pKa = 3.13
Molecular weight: 33.37 kDa
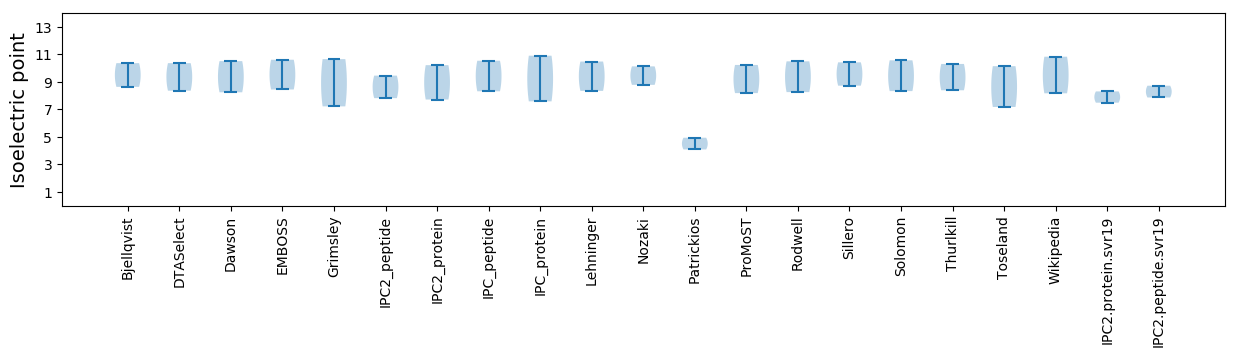
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L7UZA8|L7UZA8_9VIRU Replication associated protein OS=Cyanoramphus nest associated circular K DNA virus OX=1282444 PE=4 SV=1
MM1 pKa = 7.77PRR3 pKa = 11.84RR4 pKa = 11.84TYY6 pKa = 10.5GGRR9 pKa = 11.84SVRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84GSVRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84APMRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84VHH29 pKa = 6.11HH30 pKa = 5.88PRR32 pKa = 11.84IGRR35 pKa = 11.84GFSQYY40 pKa = 9.76VASVFKK46 pKa = 10.63TEE48 pKa = 4.15EE49 pKa = 3.74VSAGIVVAGGTSFSAFAPVSIVSMASAAASPANAVGYY86 pKa = 10.03AFQFRR91 pKa = 11.84IYY93 pKa = 10.29DD94 pKa = 3.42ISNPTSGVPSDD105 pKa = 4.09PSFITYY111 pKa = 9.35AQIYY115 pKa = 9.75DD116 pKa = 3.47QFFLTKK122 pKa = 9.09ATLVLRR128 pKa = 11.84PPLGIRR134 pKa = 11.84SFPQTVVTNAATGQIPVAQYY154 pKa = 10.54LPSTSRR160 pKa = 11.84HH161 pKa = 4.71LSYY164 pKa = 10.67IEE166 pKa = 4.4RR167 pKa = 11.84DD168 pKa = 3.16SSLALTQPLPANGDD182 pKa = 3.37GTFANRR188 pKa = 11.84ARR190 pKa = 11.84YY191 pKa = 8.21GAKK194 pKa = 9.1QHH196 pKa = 5.85SAYY199 pKa = 10.27GPIVRR204 pKa = 11.84TFWPRR209 pKa = 11.84YY210 pKa = 9.44LGAAYY215 pKa = 10.44NGADD219 pKa = 3.82YY220 pKa = 10.18TSSEE224 pKa = 4.28YY225 pKa = 11.15VIGNRR230 pKa = 11.84WLTQNRR236 pKa = 11.84SVAVGGYY243 pKa = 9.27IVLLFAYY250 pKa = 9.62NGYY253 pKa = 10.25GSAGGGPSTGAQPIINYY270 pKa = 9.5DD271 pKa = 3.58VEE273 pKa = 4.99VYY275 pKa = 7.45WHH277 pKa = 5.75VKK279 pKa = 9.12FRR281 pKa = 11.84YY282 pKa = 7.3TVYY285 pKa = 11.0GG286 pKa = 3.51
MM1 pKa = 7.77PRR3 pKa = 11.84RR4 pKa = 11.84TYY6 pKa = 10.5GGRR9 pKa = 11.84SVRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84GSVRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84APMRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84VHH29 pKa = 6.11HH30 pKa = 5.88PRR32 pKa = 11.84IGRR35 pKa = 11.84GFSQYY40 pKa = 9.76VASVFKK46 pKa = 10.63TEE48 pKa = 4.15EE49 pKa = 3.74VSAGIVVAGGTSFSAFAPVSIVSMASAAASPANAVGYY86 pKa = 10.03AFQFRR91 pKa = 11.84IYY93 pKa = 10.29DD94 pKa = 3.42ISNPTSGVPSDD105 pKa = 4.09PSFITYY111 pKa = 9.35AQIYY115 pKa = 9.75DD116 pKa = 3.47QFFLTKK122 pKa = 9.09ATLVLRR128 pKa = 11.84PPLGIRR134 pKa = 11.84SFPQTVVTNAATGQIPVAQYY154 pKa = 10.54LPSTSRR160 pKa = 11.84HH161 pKa = 4.71LSYY164 pKa = 10.67IEE166 pKa = 4.4RR167 pKa = 11.84DD168 pKa = 3.16SSLALTQPLPANGDD182 pKa = 3.37GTFANRR188 pKa = 11.84ARR190 pKa = 11.84YY191 pKa = 8.21GAKK194 pKa = 9.1QHH196 pKa = 5.85SAYY199 pKa = 10.27GPIVRR204 pKa = 11.84TFWPRR209 pKa = 11.84YY210 pKa = 9.44LGAAYY215 pKa = 10.44NGADD219 pKa = 3.82YY220 pKa = 10.18TSSEE224 pKa = 4.28YY225 pKa = 11.15VIGNRR230 pKa = 11.84WLTQNRR236 pKa = 11.84SVAVGGYY243 pKa = 9.27IVLLFAYY250 pKa = 9.62NGYY253 pKa = 10.25GSAGGGPSTGAQPIINYY270 pKa = 9.5DD271 pKa = 3.58VEE273 pKa = 4.99VYY275 pKa = 7.45WHH277 pKa = 5.75VKK279 pKa = 9.12FRR281 pKa = 11.84YY282 pKa = 7.3TVYY285 pKa = 11.0GG286 pKa = 3.51
Molecular weight: 31.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
574 |
286 |
288 |
287.0 |
32.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.537 ± 1.683 | 1.568 ± 1.146 |
4.181 ± 1.268 | 4.181 ± 1.779 |
4.181 ± 0.522 | 9.059 ± 0.534 |
2.439 ± 0.505 | 5.401 ± 0.114 |
2.962 ± 1.143 | 4.355 ± 0.139 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.697 ± 0.257 | 2.787 ± 0.518 |
5.226 ± 1.036 | 4.53 ± 0.5 |
9.582 ± 0.103 | 7.666 ± 1.298 |
7.317 ± 0.748 | 6.62 ± 1.551 |
2.091 ± 0.762 | 6.62 ± 0.528 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
