
Human respiratory syncytial virus B (strain B1)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Pneumoviridae; Orthopneumovirus; Human orthopneumovirus; Human respiratory syncytial virus B
Average proteome isoelectric point is 7.26
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
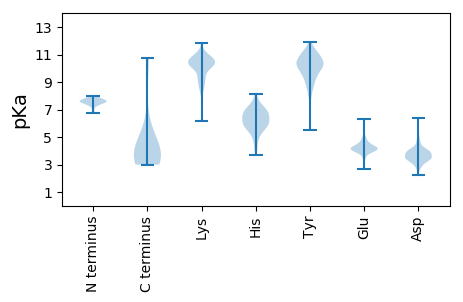
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>sp|O42083|NS1_HRSVB Non-structural protein 1 OS=Human respiratory syncytial virus B (strain B1) OX=79692 GN=1C PE=3 SV=1
MM1 pKa = 7.59EE2 pKa = 4.71KK3 pKa = 10.15FAPEE7 pKa = 3.71FHH9 pKa = 7.46GEE11 pKa = 3.97DD12 pKa = 3.96ANNKK16 pKa = 6.8ATKK19 pKa = 9.89FLEE22 pKa = 4.49SIKK25 pKa = 11.04GKK27 pKa = 9.68FASSKK32 pKa = 10.59DD33 pKa = 3.64PKK35 pKa = 10.63KK36 pKa = 10.0KK37 pKa = 10.5DD38 pKa = 3.34SIISVNSIDD47 pKa = 4.07IEE49 pKa = 4.43VTKK52 pKa = 10.35EE53 pKa = 3.85SPITSGTNIINPTSEE68 pKa = 4.3ADD70 pKa = 3.49STPEE74 pKa = 3.56TKK76 pKa = 10.56ANYY79 pKa = 7.9PRR81 pKa = 11.84KK82 pKa = 9.07PLVSFKK88 pKa = 11.1EE89 pKa = 4.01DD90 pKa = 3.61LTPSDD95 pKa = 3.85NPFSKK100 pKa = 10.44LYY102 pKa = 10.59KK103 pKa = 9.24EE104 pKa = 4.59TIEE107 pKa = 4.06TFDD110 pKa = 3.63NNEE113 pKa = 3.96EE114 pKa = 4.04EE115 pKa = 4.21SSYY118 pKa = 11.27SYY120 pKa = 11.54EE121 pKa = 4.37EE122 pKa = 5.29INDD125 pKa = 3.58QTNDD129 pKa = 3.44NITARR134 pKa = 11.84LDD136 pKa = 4.15RR137 pKa = 11.84IDD139 pKa = 4.16EE140 pKa = 4.3KK141 pKa = 11.17LSEE144 pKa = 4.26ILGMLHH150 pKa = 5.8TLVVASAGPTSARR163 pKa = 11.84DD164 pKa = 4.03GIRR167 pKa = 11.84DD168 pKa = 3.39AMVGLRR174 pKa = 11.84EE175 pKa = 3.96EE176 pKa = 4.45MIEE179 pKa = 4.21KK180 pKa = 9.97IRR182 pKa = 11.84AEE184 pKa = 3.8ALMTNDD190 pKa = 3.82RR191 pKa = 11.84LEE193 pKa = 4.27AMARR197 pKa = 11.84LRR199 pKa = 11.84NEE201 pKa = 4.35EE202 pKa = 4.19SEE204 pKa = 4.95KK205 pKa = 9.88MAKK208 pKa = 8.76DD209 pKa = 3.31TSDD212 pKa = 3.83EE213 pKa = 4.31VPLNPTSKK221 pKa = 10.69KK222 pKa = 10.68LSDD225 pKa = 3.9LLEE228 pKa = 5.54DD229 pKa = 4.0NDD231 pKa = 4.54SDD233 pKa = 5.0NDD235 pKa = 3.91LSLDD239 pKa = 3.79DD240 pKa = 4.38FF241 pKa = 5.17
MM1 pKa = 7.59EE2 pKa = 4.71KK3 pKa = 10.15FAPEE7 pKa = 3.71FHH9 pKa = 7.46GEE11 pKa = 3.97DD12 pKa = 3.96ANNKK16 pKa = 6.8ATKK19 pKa = 9.89FLEE22 pKa = 4.49SIKK25 pKa = 11.04GKK27 pKa = 9.68FASSKK32 pKa = 10.59DD33 pKa = 3.64PKK35 pKa = 10.63KK36 pKa = 10.0KK37 pKa = 10.5DD38 pKa = 3.34SIISVNSIDD47 pKa = 4.07IEE49 pKa = 4.43VTKK52 pKa = 10.35EE53 pKa = 3.85SPITSGTNIINPTSEE68 pKa = 4.3ADD70 pKa = 3.49STPEE74 pKa = 3.56TKK76 pKa = 10.56ANYY79 pKa = 7.9PRR81 pKa = 11.84KK82 pKa = 9.07PLVSFKK88 pKa = 11.1EE89 pKa = 4.01DD90 pKa = 3.61LTPSDD95 pKa = 3.85NPFSKK100 pKa = 10.44LYY102 pKa = 10.59KK103 pKa = 9.24EE104 pKa = 4.59TIEE107 pKa = 4.06TFDD110 pKa = 3.63NNEE113 pKa = 3.96EE114 pKa = 4.04EE115 pKa = 4.21SSYY118 pKa = 11.27SYY120 pKa = 11.54EE121 pKa = 4.37EE122 pKa = 5.29INDD125 pKa = 3.58QTNDD129 pKa = 3.44NITARR134 pKa = 11.84LDD136 pKa = 4.15RR137 pKa = 11.84IDD139 pKa = 4.16EE140 pKa = 4.3KK141 pKa = 11.17LSEE144 pKa = 4.26ILGMLHH150 pKa = 5.8TLVVASAGPTSARR163 pKa = 11.84DD164 pKa = 4.03GIRR167 pKa = 11.84DD168 pKa = 3.39AMVGLRR174 pKa = 11.84EE175 pKa = 3.96EE176 pKa = 4.45MIEE179 pKa = 4.21KK180 pKa = 9.97IRR182 pKa = 11.84AEE184 pKa = 3.8ALMTNDD190 pKa = 3.82RR191 pKa = 11.84LEE193 pKa = 4.27AMARR197 pKa = 11.84LRR199 pKa = 11.84NEE201 pKa = 4.35EE202 pKa = 4.19SEE204 pKa = 4.95KK205 pKa = 9.88MAKK208 pKa = 8.76DD209 pKa = 3.31TSDD212 pKa = 3.83EE213 pKa = 4.31VPLNPTSKK221 pKa = 10.69KK222 pKa = 10.68LSDD225 pKa = 3.9LLEE228 pKa = 5.54DD229 pKa = 4.0NDD231 pKa = 4.54SDD233 pKa = 5.0NDD235 pKa = 3.91LSLDD239 pKa = 3.79DD240 pKa = 4.38FF241 pKa = 5.17
Molecular weight: 26.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|O36634|FUS_HRSVB Fusion glycoprotein F0 OS=Human respiratory syncytial virus B (strain B1) OX=79692 GN=F PE=1 SV=1
MM1 pKa = 7.61SKK3 pKa = 10.42HH4 pKa = 5.73KK5 pKa = 10.15NQRR8 pKa = 11.84TARR11 pKa = 11.84TLEE14 pKa = 4.25KK15 pKa = 9.96TWDD18 pKa = 3.85TLNHH22 pKa = 6.82LIVISSCLYY31 pKa = 10.14RR32 pKa = 11.84LNLKK36 pKa = 10.43SIAQIALSVLAMIISTSLIIAAIIFIISANHH67 pKa = 6.21KK68 pKa = 8.75VTLTTVTVQTIKK80 pKa = 10.85NHH82 pKa = 4.47TEE84 pKa = 3.91KK85 pKa = 11.17NITTYY90 pKa = 9.32LTQVPPEE97 pKa = 4.09RR98 pKa = 11.84VSSSKK103 pKa = 10.67QPTTTSPIHH112 pKa = 5.99TNSATTSPNTKK123 pKa = 10.0SEE125 pKa = 4.22THH127 pKa = 5.91HH128 pKa = 5.9TTAQTKK134 pKa = 10.05GRR136 pKa = 11.84TTTSTQTNKK145 pKa = 10.25PSTKK149 pKa = 9.48PRR151 pKa = 11.84LKK153 pKa = 10.72NPPKK157 pKa = 10.29KK158 pKa = 10.31PKK160 pKa = 9.94DD161 pKa = 3.65DD162 pKa = 3.53YY163 pKa = 11.35HH164 pKa = 8.1FEE166 pKa = 4.02VFNFVPCSICGNNQLCKK183 pKa = 10.21SICKK187 pKa = 8.93TIPSNKK193 pKa = 8.61PKK195 pKa = 10.34KK196 pKa = 10.27KK197 pKa = 8.93PTIKK201 pKa = 9.27PTNKK205 pKa = 8.47PTTKK209 pKa = 8.25TTNKK213 pKa = 9.62RR214 pKa = 11.84DD215 pKa = 3.54PKK217 pKa = 10.43TPAKK221 pKa = 6.29TTKK224 pKa = 10.47KK225 pKa = 9.01EE226 pKa = 4.13TTTNPTKK233 pKa = 10.7KK234 pKa = 8.82PTLTTTEE241 pKa = 4.53RR242 pKa = 11.84DD243 pKa = 3.41TSTSQSTVLDD253 pKa = 3.89TTTLEE258 pKa = 4.25HH259 pKa = 7.08TIQQQSLHH267 pKa = 5.77STTPEE272 pKa = 3.55NTPNSTQTPTASEE285 pKa = 4.36PSTSNSTQNTQSHH298 pKa = 5.97AA299 pKa = 3.75
MM1 pKa = 7.61SKK3 pKa = 10.42HH4 pKa = 5.73KK5 pKa = 10.15NQRR8 pKa = 11.84TARR11 pKa = 11.84TLEE14 pKa = 4.25KK15 pKa = 9.96TWDD18 pKa = 3.85TLNHH22 pKa = 6.82LIVISSCLYY31 pKa = 10.14RR32 pKa = 11.84LNLKK36 pKa = 10.43SIAQIALSVLAMIISTSLIIAAIIFIISANHH67 pKa = 6.21KK68 pKa = 8.75VTLTTVTVQTIKK80 pKa = 10.85NHH82 pKa = 4.47TEE84 pKa = 3.91KK85 pKa = 11.17NITTYY90 pKa = 9.32LTQVPPEE97 pKa = 4.09RR98 pKa = 11.84VSSSKK103 pKa = 10.67QPTTTSPIHH112 pKa = 5.99TNSATTSPNTKK123 pKa = 10.0SEE125 pKa = 4.22THH127 pKa = 5.91HH128 pKa = 5.9TTAQTKK134 pKa = 10.05GRR136 pKa = 11.84TTTSTQTNKK145 pKa = 10.25PSTKK149 pKa = 9.48PRR151 pKa = 11.84LKK153 pKa = 10.72NPPKK157 pKa = 10.29KK158 pKa = 10.31PKK160 pKa = 9.94DD161 pKa = 3.65DD162 pKa = 3.53YY163 pKa = 11.35HH164 pKa = 8.1FEE166 pKa = 4.02VFNFVPCSICGNNQLCKK183 pKa = 10.21SICKK187 pKa = 8.93TIPSNKK193 pKa = 8.61PKK195 pKa = 10.34KK196 pKa = 10.27KK197 pKa = 8.93PTIKK201 pKa = 9.27PTNKK205 pKa = 8.47PTTKK209 pKa = 8.25TTNKK213 pKa = 9.62RR214 pKa = 11.84DD215 pKa = 3.54PKK217 pKa = 10.43TPAKK221 pKa = 6.29TTKK224 pKa = 10.47KK225 pKa = 9.01EE226 pKa = 4.13TTTNPTKK233 pKa = 10.7KK234 pKa = 8.82PTLTTTEE241 pKa = 4.53RR242 pKa = 11.84DD243 pKa = 3.41TSTSQSTVLDD253 pKa = 3.89TTTLEE258 pKa = 4.25HH259 pKa = 7.08TIQQQSLHH267 pKa = 5.77STTPEE272 pKa = 3.55NTPNSTQTPTASEE285 pKa = 4.36PSTSNSTQNTQSHH298 pKa = 5.97AA299 pKa = 3.75
Molecular weight: 32.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4792 |
65 |
2166 |
399.3 |
45.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.111 ± 0.59 | 1.732 ± 0.204 |
4.633 ± 0.523 | 5.071 ± 0.561 |
3.443 ± 0.487 | 3.652 ± 0.577 |
2.504 ± 0.339 | 9.015 ± 0.478 |
8.076 ± 0.445 | 10.538 ± 1.176 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.295 ± 0.286 | 7.846 ± 0.334 |
3.902 ± 0.763 | 3.13 ± 0.353 |
3.339 ± 0.275 | 8.598 ± 0.427 |
8.598 ± 2.268 | 4.883 ± 0.456 |
0.772 ± 0.227 | 3.861 ± 0.632 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
