
Gemycircularvirus C1c
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus humas3; Human associated gemykibivirus 3
Average proteome isoelectric point is 9.31
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0R7I4J2|A0A0R7I4J2_9VIRU Capsid protein OS=Gemycircularvirus C1c OX=1673681 GN=Cap PE=4 SV=1
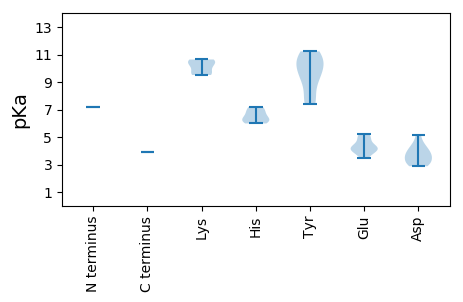
MM1 pKa = 7.18FFFVEE6 pKa = 5.19GYY8 pKa = 8.98VAFYY12 pKa = 11.23VMAYY16 pKa = 7.66AHH18 pKa = 6.23RR19 pKa = 11.84RR20 pKa = 11.84FRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84FARR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84PTFARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84VPRR40 pKa = 11.84RR41 pKa = 11.84SYY43 pKa = 9.71GRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SSGRR52 pKa = 11.84SVRR55 pKa = 11.84SIRR58 pKa = 11.84NIAARR63 pKa = 11.84KK64 pKa = 9.55CRR66 pKa = 11.84DD67 pKa = 3.06NMLGVPISTDD77 pKa = 3.61FQQQNPGPLTMFGDD91 pKa = 4.18TSHH94 pKa = 6.97GYY96 pKa = 10.21LFSPTARR103 pKa = 11.84RR104 pKa = 11.84VGSLDD109 pKa = 2.88GSLPRR114 pKa = 11.84GIHH117 pKa = 5.99ANSAEE122 pKa = 4.02RR123 pKa = 11.84WKK125 pKa = 9.51TRR127 pKa = 11.84CFVKK131 pKa = 10.58GLQEE135 pKa = 4.15TVEE138 pKa = 4.09LQPNNGAGWLWRR150 pKa = 11.84RR151 pKa = 11.84VVFSCIGLLEE161 pKa = 4.23EE162 pKa = 5.0FPTNSVAAPDD172 pKa = 3.55STRR175 pKa = 11.84GYY177 pKa = 10.92GRR179 pKa = 11.84AIWNMRR185 pKa = 11.84DD186 pKa = 3.05GSAAATPLYY195 pKa = 11.1NEE197 pKa = 3.87MANYY201 pKa = 9.99LFEE204 pKa = 4.34GTRR207 pKa = 11.84LRR209 pKa = 11.84DD210 pKa = 3.54WINPFTAKK218 pKa = 10.26LDD220 pKa = 3.75RR221 pKa = 11.84SIVTVHH227 pKa = 6.71SDD229 pKa = 3.16TTRR232 pKa = 11.84TIQSNNPNGTYY243 pKa = 10.26KK244 pKa = 10.18IFKK247 pKa = 9.71RR248 pKa = 11.84YY249 pKa = 9.47YY250 pKa = 7.6PVNRR254 pKa = 11.84GIVYY258 pKa = 10.49ADD260 pKa = 4.25DD261 pKa = 4.07EE262 pKa = 4.91SGSGSKK268 pKa = 10.59NDD270 pKa = 3.06AHH272 pKa = 6.36YY273 pKa = 10.8AAGTTKK279 pKa = 10.64GNSGDD284 pKa = 4.59LFVLDD289 pKa = 5.15LFNAINDD296 pKa = 3.73SSEE299 pKa = 3.49NTMNFGVHH307 pKa = 6.16ARR309 pKa = 11.84YY310 pKa = 8.98YY311 pKa = 7.43WHH313 pKa = 7.21EE314 pKa = 4.33GSGEE318 pKa = 3.88
MM1 pKa = 7.18FFFVEE6 pKa = 5.19GYY8 pKa = 8.98VAFYY12 pKa = 11.23VMAYY16 pKa = 7.66AHH18 pKa = 6.23RR19 pKa = 11.84RR20 pKa = 11.84FRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84FARR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84PTFARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84VPRR40 pKa = 11.84RR41 pKa = 11.84SYY43 pKa = 9.71GRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SSGRR52 pKa = 11.84SVRR55 pKa = 11.84SIRR58 pKa = 11.84NIAARR63 pKa = 11.84KK64 pKa = 9.55CRR66 pKa = 11.84DD67 pKa = 3.06NMLGVPISTDD77 pKa = 3.61FQQQNPGPLTMFGDD91 pKa = 4.18TSHH94 pKa = 6.97GYY96 pKa = 10.21LFSPTARR103 pKa = 11.84RR104 pKa = 11.84VGSLDD109 pKa = 2.88GSLPRR114 pKa = 11.84GIHH117 pKa = 5.99ANSAEE122 pKa = 4.02RR123 pKa = 11.84WKK125 pKa = 9.51TRR127 pKa = 11.84CFVKK131 pKa = 10.58GLQEE135 pKa = 4.15TVEE138 pKa = 4.09LQPNNGAGWLWRR150 pKa = 11.84RR151 pKa = 11.84VVFSCIGLLEE161 pKa = 4.23EE162 pKa = 5.0FPTNSVAAPDD172 pKa = 3.55STRR175 pKa = 11.84GYY177 pKa = 10.92GRR179 pKa = 11.84AIWNMRR185 pKa = 11.84DD186 pKa = 3.05GSAAATPLYY195 pKa = 11.1NEE197 pKa = 3.87MANYY201 pKa = 9.99LFEE204 pKa = 4.34GTRR207 pKa = 11.84LRR209 pKa = 11.84DD210 pKa = 3.54WINPFTAKK218 pKa = 10.26LDD220 pKa = 3.75RR221 pKa = 11.84SIVTVHH227 pKa = 6.71SDD229 pKa = 3.16TTRR232 pKa = 11.84TIQSNNPNGTYY243 pKa = 10.26KK244 pKa = 10.18IFKK247 pKa = 9.71RR248 pKa = 11.84YY249 pKa = 9.47YY250 pKa = 7.6PVNRR254 pKa = 11.84GIVYY258 pKa = 10.49ADD260 pKa = 4.25DD261 pKa = 4.07EE262 pKa = 4.91SGSGSKK268 pKa = 10.59NDD270 pKa = 3.06AHH272 pKa = 6.36YY273 pKa = 10.8AAGTTKK279 pKa = 10.64GNSGDD284 pKa = 4.59LFVLDD289 pKa = 5.15LFNAINDD296 pKa = 3.73SSEE299 pKa = 3.49NTMNFGVHH307 pKa = 6.16ARR309 pKa = 11.84YY310 pKa = 8.98YY311 pKa = 7.43WHH313 pKa = 7.21EE314 pKa = 4.33GSGEE318 pKa = 3.88
Molecular weight: 36.28 kDa
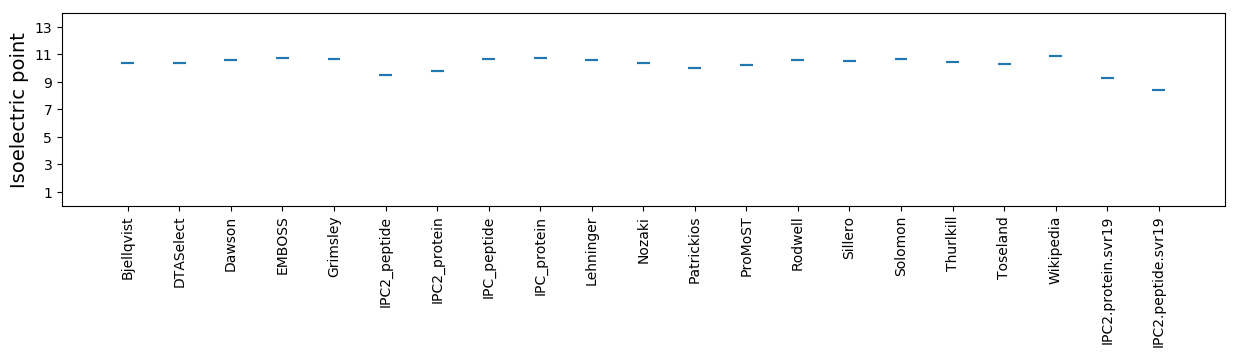
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0R7I4J2|A0A0R7I4J2_9VIRU Capsid protein OS=Gemycircularvirus C1c OX=1673681 GN=Cap PE=4 SV=1
MM1 pKa = 7.18FFFVEE6 pKa = 5.19GYY8 pKa = 8.98VAFYY12 pKa = 11.23VMAYY16 pKa = 7.66AHH18 pKa = 6.23RR19 pKa = 11.84RR20 pKa = 11.84FRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84FARR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84PTFARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84VPRR40 pKa = 11.84RR41 pKa = 11.84SYY43 pKa = 9.71GRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SSGRR52 pKa = 11.84SVRR55 pKa = 11.84SIRR58 pKa = 11.84NIAARR63 pKa = 11.84KK64 pKa = 9.55CRR66 pKa = 11.84DD67 pKa = 3.06NMLGVPISTDD77 pKa = 3.61FQQQNPGPLTMFGDD91 pKa = 4.18TSHH94 pKa = 6.97GYY96 pKa = 10.21LFSPTARR103 pKa = 11.84RR104 pKa = 11.84VGSLDD109 pKa = 2.88GSLPRR114 pKa = 11.84GIHH117 pKa = 5.99ANSAEE122 pKa = 4.02RR123 pKa = 11.84WKK125 pKa = 9.51TRR127 pKa = 11.84CFVKK131 pKa = 10.58GLQEE135 pKa = 4.15TVEE138 pKa = 4.09LQPNNGAGWLWRR150 pKa = 11.84RR151 pKa = 11.84VVFSCIGLLEE161 pKa = 4.23EE162 pKa = 5.0FPTNSVAAPDD172 pKa = 3.55STRR175 pKa = 11.84GYY177 pKa = 10.92GRR179 pKa = 11.84AIWNMRR185 pKa = 11.84DD186 pKa = 3.05GSAAATPLYY195 pKa = 11.1NEE197 pKa = 3.87MANYY201 pKa = 9.99LFEE204 pKa = 4.34GTRR207 pKa = 11.84LRR209 pKa = 11.84DD210 pKa = 3.54WINPFTAKK218 pKa = 10.26LDD220 pKa = 3.75RR221 pKa = 11.84SIVTVHH227 pKa = 6.71SDD229 pKa = 3.16TTRR232 pKa = 11.84TIQSNNPNGTYY243 pKa = 10.26KK244 pKa = 10.18IFKK247 pKa = 9.71RR248 pKa = 11.84YY249 pKa = 9.47YY250 pKa = 7.6PVNRR254 pKa = 11.84GIVYY258 pKa = 10.49ADD260 pKa = 4.25DD261 pKa = 4.07EE262 pKa = 4.91SGSGSKK268 pKa = 10.59NDD270 pKa = 3.06AHH272 pKa = 6.36YY273 pKa = 10.8AAGTTKK279 pKa = 10.64GNSGDD284 pKa = 4.59LFVLDD289 pKa = 5.15LFNAINDD296 pKa = 3.73SSEE299 pKa = 3.49NTMNFGVHH307 pKa = 6.16ARR309 pKa = 11.84YY310 pKa = 8.98YY311 pKa = 7.43WHH313 pKa = 7.21EE314 pKa = 4.33GSGEE318 pKa = 3.88
MM1 pKa = 7.18FFFVEE6 pKa = 5.19GYY8 pKa = 8.98VAFYY12 pKa = 11.23VMAYY16 pKa = 7.66AHH18 pKa = 6.23RR19 pKa = 11.84RR20 pKa = 11.84FRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84FARR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84PTFARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84VPRR40 pKa = 11.84RR41 pKa = 11.84SYY43 pKa = 9.71GRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SSGRR52 pKa = 11.84SVRR55 pKa = 11.84SIRR58 pKa = 11.84NIAARR63 pKa = 11.84KK64 pKa = 9.55CRR66 pKa = 11.84DD67 pKa = 3.06NMLGVPISTDD77 pKa = 3.61FQQQNPGPLTMFGDD91 pKa = 4.18TSHH94 pKa = 6.97GYY96 pKa = 10.21LFSPTARR103 pKa = 11.84RR104 pKa = 11.84VGSLDD109 pKa = 2.88GSLPRR114 pKa = 11.84GIHH117 pKa = 5.99ANSAEE122 pKa = 4.02RR123 pKa = 11.84WKK125 pKa = 9.51TRR127 pKa = 11.84CFVKK131 pKa = 10.58GLQEE135 pKa = 4.15TVEE138 pKa = 4.09LQPNNGAGWLWRR150 pKa = 11.84RR151 pKa = 11.84VVFSCIGLLEE161 pKa = 4.23EE162 pKa = 5.0FPTNSVAAPDD172 pKa = 3.55STRR175 pKa = 11.84GYY177 pKa = 10.92GRR179 pKa = 11.84AIWNMRR185 pKa = 11.84DD186 pKa = 3.05GSAAATPLYY195 pKa = 11.1NEE197 pKa = 3.87MANYY201 pKa = 9.99LFEE204 pKa = 4.34GTRR207 pKa = 11.84LRR209 pKa = 11.84DD210 pKa = 3.54WINPFTAKK218 pKa = 10.26LDD220 pKa = 3.75RR221 pKa = 11.84SIVTVHH227 pKa = 6.71SDD229 pKa = 3.16TTRR232 pKa = 11.84TIQSNNPNGTYY243 pKa = 10.26KK244 pKa = 10.18IFKK247 pKa = 9.71RR248 pKa = 11.84YY249 pKa = 9.47YY250 pKa = 7.6PVNRR254 pKa = 11.84GIVYY258 pKa = 10.49ADD260 pKa = 4.25DD261 pKa = 4.07EE262 pKa = 4.91SGSGSKK268 pKa = 10.59NDD270 pKa = 3.06AHH272 pKa = 6.36YY273 pKa = 10.8AAGTTKK279 pKa = 10.64GNSGDD284 pKa = 4.59LFVLDD289 pKa = 5.15LFNAINDD296 pKa = 3.73SSEE299 pKa = 3.49NTMNFGVHH307 pKa = 6.16ARR309 pKa = 11.84YY310 pKa = 8.98YY311 pKa = 7.43WHH313 pKa = 7.21EE314 pKa = 4.33GSGEE318 pKa = 3.88
Molecular weight: 36.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
318 |
318 |
318 |
318.0 |
36.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.862 ± 0.0 | 0.943 ± 0.0 |
4.717 ± 0.0 | 3.774 ± 0.0 |
5.975 ± 0.0 | 8.805 ± 0.0 |
2.201 ± 0.0 | 3.774 ± 0.0 |
2.516 ± 0.0 | 5.346 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.201 ± 0.0 | 6.604 ± 0.0 |
4.403 ± 0.0 | 1.887 ± 0.0 |
12.579 ± 0.0 | 7.862 ± 0.0 |
6.289 ± 0.0 | 5.66 ± 0.0 |
1.887 ± 0.0 | 4.717 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
