
Planococcus salinus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Planococcaceae; Planococcus
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
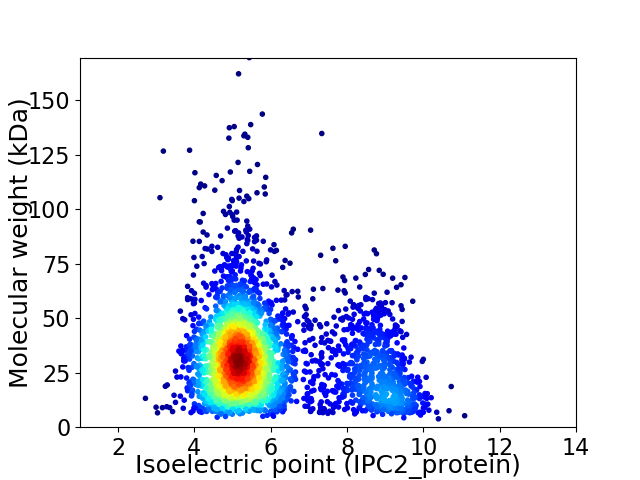
Virtual 2D-PAGE plot for 3216 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3M8P7P5|A0A3M8P7P5_9BACL Class I SAM-dependent methyltransferase OS=Planococcus salinus OX=1848460 GN=EEX84_07040 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.51NSFKK6 pKa = 11.13VIVLFLCSILILSGCAGFQTSNSDD30 pKa = 3.34EE31 pKa = 4.44GNTNEE36 pKa = 4.98DD37 pKa = 3.19GKK39 pKa = 9.15TVVDD43 pKa = 6.07FWSFWGSEE51 pKa = 3.89LRR53 pKa = 11.84RR54 pKa = 11.84PIIDD58 pKa = 4.66KK59 pKa = 10.64IVADD63 pKa = 5.32FNASQDD69 pKa = 3.87EE70 pKa = 4.5IEE72 pKa = 4.52VKK74 pKa = 8.92HH75 pKa = 6.1TYY77 pKa = 10.39SPWGDD82 pKa = 2.39IWTKK86 pKa = 10.4EE87 pKa = 3.83LAAIAAGNPPDD98 pKa = 5.08VVINDD103 pKa = 3.42INATAIRR110 pKa = 11.84GQEE113 pKa = 3.85NQAMNLSEE121 pKa = 5.21FYY123 pKa = 10.78DD124 pKa = 3.73DD125 pKa = 4.56SFSEE129 pKa = 4.15KK130 pKa = 10.25FYY132 pKa = 10.82PEE134 pKa = 3.91LWNATLYY141 pKa = 10.99EE142 pKa = 4.5GDD144 pKa = 3.69SYY146 pKa = 11.53GIPFNTDD153 pKa = 2.29TRR155 pKa = 11.84VLYY158 pKa = 10.02YY159 pKa = 10.9NKK161 pKa = 10.36DD162 pKa = 3.08AFEE165 pKa = 4.24EE166 pKa = 4.32VGLDD170 pKa = 3.62PEE172 pKa = 5.19DD173 pKa = 4.58PPEE176 pKa = 3.91TWEE179 pKa = 4.01EE180 pKa = 4.13LEE182 pKa = 4.39EE183 pKa = 4.07YY184 pKa = 10.1AAKK187 pKa = 10.6LDD189 pKa = 3.83VKK191 pKa = 10.89NGEE194 pKa = 4.17SYY196 pKa = 11.31DD197 pKa = 3.68RR198 pKa = 11.84LGFYY202 pKa = 9.8PLWGVGYY209 pKa = 9.73DD210 pKa = 3.47VWLLNANGEE219 pKa = 4.42NYY221 pKa = 10.22FGADD225 pKa = 3.34NNVDD229 pKa = 3.65TVNVNTEE236 pKa = 4.13TNVEE240 pKa = 4.0VLNWLKK246 pKa = 10.49SWRR249 pKa = 11.84DD250 pKa = 3.51RR251 pKa = 11.84IGEE254 pKa = 4.17DD255 pKa = 3.93VVNSYY260 pKa = 9.05QAQIDD265 pKa = 4.15SQQGNPFFSGDD276 pKa = 3.37LAMLVQQPTFYY287 pKa = 10.45TQIRR291 pKa = 11.84DD292 pKa = 3.57YY293 pKa = 11.8AEE295 pKa = 4.28DD296 pKa = 4.04LNFGVAQLPEE306 pKa = 4.17YY307 pKa = 10.58EE308 pKa = 4.82PGNGHH313 pKa = 5.81TSWGGGFVAEE323 pKa = 4.38IPEE326 pKa = 4.59GSSDD330 pKa = 5.05PEE332 pKa = 3.9AAWAFIEE339 pKa = 4.48YY340 pKa = 8.7LTGPEE345 pKa = 3.93AQEE348 pKa = 3.67YY349 pKa = 8.05WAVQNFDD356 pKa = 3.02NVANIEE362 pKa = 4.05AAEE365 pKa = 4.11AAAQSDD371 pKa = 4.21EE372 pKa = 4.17FTEE375 pKa = 4.46EE376 pKa = 3.91GQMVYY381 pKa = 10.43QMAVDD386 pKa = 3.64SMEE389 pKa = 4.18NTLLTPAPVEE399 pKa = 3.86APGFYY404 pKa = 10.6NYY406 pKa = 9.96INPKK410 pKa = 9.44VDD412 pKa = 3.32EE413 pKa = 4.77FFLGSLTAEE422 pKa = 4.11EE423 pKa = 4.97ALAQAQADD431 pKa = 3.92VEE433 pKa = 4.46ALIEE437 pKa = 4.49SNQQ440 pKa = 3.22
MM1 pKa = 7.47KK2 pKa = 10.51NSFKK6 pKa = 11.13VIVLFLCSILILSGCAGFQTSNSDD30 pKa = 3.34EE31 pKa = 4.44GNTNEE36 pKa = 4.98DD37 pKa = 3.19GKK39 pKa = 9.15TVVDD43 pKa = 6.07FWSFWGSEE51 pKa = 3.89LRR53 pKa = 11.84RR54 pKa = 11.84PIIDD58 pKa = 4.66KK59 pKa = 10.64IVADD63 pKa = 5.32FNASQDD69 pKa = 3.87EE70 pKa = 4.5IEE72 pKa = 4.52VKK74 pKa = 8.92HH75 pKa = 6.1TYY77 pKa = 10.39SPWGDD82 pKa = 2.39IWTKK86 pKa = 10.4EE87 pKa = 3.83LAAIAAGNPPDD98 pKa = 5.08VVINDD103 pKa = 3.42INATAIRR110 pKa = 11.84GQEE113 pKa = 3.85NQAMNLSEE121 pKa = 5.21FYY123 pKa = 10.78DD124 pKa = 3.73DD125 pKa = 4.56SFSEE129 pKa = 4.15KK130 pKa = 10.25FYY132 pKa = 10.82PEE134 pKa = 3.91LWNATLYY141 pKa = 10.99EE142 pKa = 4.5GDD144 pKa = 3.69SYY146 pKa = 11.53GIPFNTDD153 pKa = 2.29TRR155 pKa = 11.84VLYY158 pKa = 10.02YY159 pKa = 10.9NKK161 pKa = 10.36DD162 pKa = 3.08AFEE165 pKa = 4.24EE166 pKa = 4.32VGLDD170 pKa = 3.62PEE172 pKa = 5.19DD173 pKa = 4.58PPEE176 pKa = 3.91TWEE179 pKa = 4.01EE180 pKa = 4.13LEE182 pKa = 4.39EE183 pKa = 4.07YY184 pKa = 10.1AAKK187 pKa = 10.6LDD189 pKa = 3.83VKK191 pKa = 10.89NGEE194 pKa = 4.17SYY196 pKa = 11.31DD197 pKa = 3.68RR198 pKa = 11.84LGFYY202 pKa = 9.8PLWGVGYY209 pKa = 9.73DD210 pKa = 3.47VWLLNANGEE219 pKa = 4.42NYY221 pKa = 10.22FGADD225 pKa = 3.34NNVDD229 pKa = 3.65TVNVNTEE236 pKa = 4.13TNVEE240 pKa = 4.0VLNWLKK246 pKa = 10.49SWRR249 pKa = 11.84DD250 pKa = 3.51RR251 pKa = 11.84IGEE254 pKa = 4.17DD255 pKa = 3.93VVNSYY260 pKa = 9.05QAQIDD265 pKa = 4.15SQQGNPFFSGDD276 pKa = 3.37LAMLVQQPTFYY287 pKa = 10.45TQIRR291 pKa = 11.84DD292 pKa = 3.57YY293 pKa = 11.8AEE295 pKa = 4.28DD296 pKa = 4.04LNFGVAQLPEE306 pKa = 4.17YY307 pKa = 10.58EE308 pKa = 4.82PGNGHH313 pKa = 5.81TSWGGGFVAEE323 pKa = 4.38IPEE326 pKa = 4.59GSSDD330 pKa = 5.05PEE332 pKa = 3.9AAWAFIEE339 pKa = 4.48YY340 pKa = 8.7LTGPEE345 pKa = 3.93AQEE348 pKa = 3.67YY349 pKa = 8.05WAVQNFDD356 pKa = 3.02NVANIEE362 pKa = 4.05AAEE365 pKa = 4.11AAAQSDD371 pKa = 4.21EE372 pKa = 4.17FTEE375 pKa = 4.46EE376 pKa = 3.91GQMVYY381 pKa = 10.43QMAVDD386 pKa = 3.64SMEE389 pKa = 4.18NTLLTPAPVEE399 pKa = 3.86APGFYY404 pKa = 10.6NYY406 pKa = 9.96INPKK410 pKa = 9.44VDD412 pKa = 3.32EE413 pKa = 4.77FFLGSLTAEE422 pKa = 4.11EE423 pKa = 4.97ALAQAQADD431 pKa = 3.92VEE433 pKa = 4.46ALIEE437 pKa = 4.49SNQQ440 pKa = 3.22
Molecular weight: 49.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M8P8K1|A0A3M8P8K1_9BACL DNA replication and repair protein RecF OS=Planococcus salinus OX=1848460 GN=recF PE=3 SV=1
MM1 pKa = 7.67SLRR4 pKa = 11.84TYY6 pKa = 10.39QPNTRR11 pKa = 11.84KK12 pKa = 9.66HH13 pKa = 5.87SKK15 pKa = 8.83VHH17 pKa = 5.7GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.71NGRR29 pKa = 11.84RR30 pKa = 11.84VIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.8GRR40 pKa = 11.84KK41 pKa = 8.75VLSAA45 pKa = 4.05
MM1 pKa = 7.67SLRR4 pKa = 11.84TYY6 pKa = 10.39QPNTRR11 pKa = 11.84KK12 pKa = 9.66HH13 pKa = 5.87SKK15 pKa = 8.83VHH17 pKa = 5.7GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.71NGRR29 pKa = 11.84RR30 pKa = 11.84VIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.8GRR40 pKa = 11.84KK41 pKa = 8.75VLSAA45 pKa = 4.05
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
962098 |
37 |
1528 |
299.2 |
33.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.111 ± 0.045 | 0.584 ± 0.013 |
5.204 ± 0.037 | 7.989 ± 0.057 |
4.523 ± 0.035 | 7.247 ± 0.037 |
2.042 ± 0.022 | 7.054 ± 0.044 |
5.944 ± 0.046 | 9.717 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.917 ± 0.021 | 3.777 ± 0.026 |
3.828 ± 0.026 | 3.765 ± 0.029 |
4.312 ± 0.031 | 5.901 ± 0.028 |
5.508 ± 0.026 | 7.327 ± 0.029 |
1.023 ± 0.017 | 3.227 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
