
Spiribacter sp. E85
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; Spiribacter; unclassified Spiribacter
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
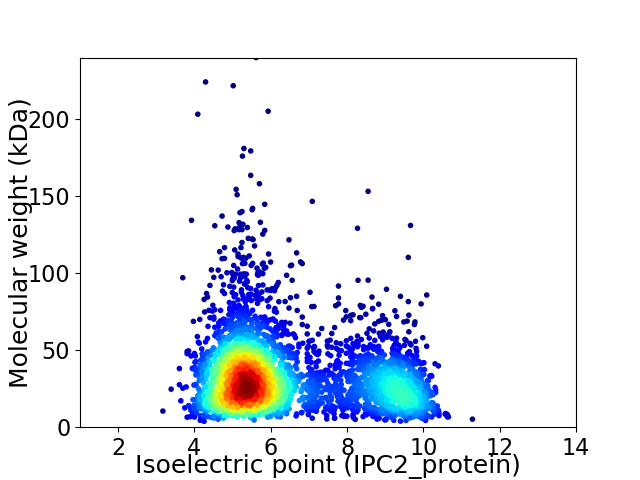
Virtual 2D-PAGE plot for 3732 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U2MX23|A0A2U2MX23_9GAMM 23S rRNA (guanosine-2'-O-)-methyltransferase RlmB OS=Spiribacter sp. E85 OX=2182432 GN=rlmB PE=3 SV=1
MM1 pKa = 6.92MKK3 pKa = 8.91KK4 pKa = 7.86TASALAVAALMGTGAASAATFQVNDD29 pKa = 3.74DD30 pKa = 4.21TTLSLYY36 pKa = 11.17GNVQIAYY43 pKa = 10.1SDD45 pKa = 3.83TNDD48 pKa = 3.14ASGDD52 pKa = 4.23SVSDD56 pKa = 3.64LQDD59 pKa = 3.12NGTTIGVAVEE69 pKa = 3.96HH70 pKa = 6.76RR71 pKa = 11.84FDD73 pKa = 3.73NGLTGFARR81 pKa = 11.84IEE83 pKa = 3.95EE84 pKa = 4.48DD85 pKa = 3.74GYY87 pKa = 11.63DD88 pKa = 3.64AVEE91 pKa = 4.19QAAEE95 pKa = 4.01TAATDD100 pKa = 3.29QAYY103 pKa = 10.2FGVRR107 pKa = 11.84GDD109 pKa = 3.61FGMVRR114 pKa = 11.84LGNADD119 pKa = 3.76SIYY122 pKa = 10.79DD123 pKa = 3.77GFVRR127 pKa = 11.84DD128 pKa = 3.46YY129 pKa = 10.88WDD131 pKa = 3.42YY132 pKa = 11.86QEE134 pKa = 5.71FLAPINGRR142 pKa = 11.84RR143 pKa = 11.84GVDD146 pKa = 2.98HH147 pKa = 7.95KK148 pKa = 11.22DD149 pKa = 3.16RR150 pKa = 11.84QLLYY154 pKa = 9.64MSPNFGGFSFGLEE167 pKa = 3.77AQINGDD173 pKa = 3.27ADD175 pKa = 3.55EE176 pKa = 4.76ATFEE180 pKa = 4.19LTQGGVNNGDD190 pKa = 3.75NEE192 pKa = 4.49TGDD195 pKa = 3.83VNGDD199 pKa = 3.49DD200 pKa = 5.0AGISLAAAAMYY211 pKa = 7.58EE212 pKa = 4.33TGGLTVSAAIDD223 pKa = 3.35QRR225 pKa = 11.84SNEE228 pKa = 4.17FVSAADD234 pKa = 3.77AGSEE238 pKa = 3.95GGPNHH243 pKa = 6.7FDD245 pKa = 3.61DD246 pKa = 4.6PVMGIVVSYY255 pKa = 10.97DD256 pKa = 3.18FGRR259 pKa = 11.84FTVDD263 pKa = 3.43GGYY266 pKa = 10.13QLDD269 pKa = 4.06AADD272 pKa = 3.75TDD274 pKa = 3.57EE275 pKa = 4.75RR276 pKa = 11.84AIGDD280 pKa = 3.71VDD282 pKa = 3.87VLSLGGRR289 pKa = 11.84FNYY292 pKa = 10.54GMGDD296 pKa = 3.54LYY298 pKa = 11.36GIVQEE303 pKa = 4.28VGYY306 pKa = 10.63DD307 pKa = 3.56EE308 pKa = 5.39EE309 pKa = 4.33NAAGNDD315 pKa = 3.33SFTEE319 pKa = 4.51VILGANYY326 pKa = 10.45NIGSNFYY333 pKa = 10.85VYY335 pKa = 10.9AEE337 pKa = 4.17YY338 pKa = 8.63ATYY341 pKa = 10.41DD342 pKa = 3.53QEE344 pKa = 4.5EE345 pKa = 4.25DD346 pKa = 3.79ANDD349 pKa = 3.5GFAIGGLYY357 pKa = 10.55SFF359 pKa = 5.56
MM1 pKa = 6.92MKK3 pKa = 8.91KK4 pKa = 7.86TASALAVAALMGTGAASAATFQVNDD29 pKa = 3.74DD30 pKa = 4.21TTLSLYY36 pKa = 11.17GNVQIAYY43 pKa = 10.1SDD45 pKa = 3.83TNDD48 pKa = 3.14ASGDD52 pKa = 4.23SVSDD56 pKa = 3.64LQDD59 pKa = 3.12NGTTIGVAVEE69 pKa = 3.96HH70 pKa = 6.76RR71 pKa = 11.84FDD73 pKa = 3.73NGLTGFARR81 pKa = 11.84IEE83 pKa = 3.95EE84 pKa = 4.48DD85 pKa = 3.74GYY87 pKa = 11.63DD88 pKa = 3.64AVEE91 pKa = 4.19QAAEE95 pKa = 4.01TAATDD100 pKa = 3.29QAYY103 pKa = 10.2FGVRR107 pKa = 11.84GDD109 pKa = 3.61FGMVRR114 pKa = 11.84LGNADD119 pKa = 3.76SIYY122 pKa = 10.79DD123 pKa = 3.77GFVRR127 pKa = 11.84DD128 pKa = 3.46YY129 pKa = 10.88WDD131 pKa = 3.42YY132 pKa = 11.86QEE134 pKa = 5.71FLAPINGRR142 pKa = 11.84RR143 pKa = 11.84GVDD146 pKa = 2.98HH147 pKa = 7.95KK148 pKa = 11.22DD149 pKa = 3.16RR150 pKa = 11.84QLLYY154 pKa = 9.64MSPNFGGFSFGLEE167 pKa = 3.77AQINGDD173 pKa = 3.27ADD175 pKa = 3.55EE176 pKa = 4.76ATFEE180 pKa = 4.19LTQGGVNNGDD190 pKa = 3.75NEE192 pKa = 4.49TGDD195 pKa = 3.83VNGDD199 pKa = 3.49DD200 pKa = 5.0AGISLAAAAMYY211 pKa = 7.58EE212 pKa = 4.33TGGLTVSAAIDD223 pKa = 3.35QRR225 pKa = 11.84SNEE228 pKa = 4.17FVSAADD234 pKa = 3.77AGSEE238 pKa = 3.95GGPNHH243 pKa = 6.7FDD245 pKa = 3.61DD246 pKa = 4.6PVMGIVVSYY255 pKa = 10.97DD256 pKa = 3.18FGRR259 pKa = 11.84FTVDD263 pKa = 3.43GGYY266 pKa = 10.13QLDD269 pKa = 4.06AADD272 pKa = 3.75TDD274 pKa = 3.57EE275 pKa = 4.75RR276 pKa = 11.84AIGDD280 pKa = 3.71VDD282 pKa = 3.87VLSLGGRR289 pKa = 11.84FNYY292 pKa = 10.54GMGDD296 pKa = 3.54LYY298 pKa = 11.36GIVQEE303 pKa = 4.28VGYY306 pKa = 10.63DD307 pKa = 3.56EE308 pKa = 5.39EE309 pKa = 4.33NAAGNDD315 pKa = 3.33SFTEE319 pKa = 4.51VILGANYY326 pKa = 10.45NIGSNFYY333 pKa = 10.85VYY335 pKa = 10.9AEE337 pKa = 4.17YY338 pKa = 8.63ATYY341 pKa = 10.41DD342 pKa = 3.53QEE344 pKa = 4.5EE345 pKa = 4.25DD346 pKa = 3.79ANDD349 pKa = 3.5GFAIGGLYY357 pKa = 10.55SFF359 pKa = 5.56
Molecular weight: 38.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U2N2X6|A0A2U2N2X6_9GAMM Probable chromosome-partitioning protein ParB OS=Spiribacter sp. E85 OX=2182432 GN=DEM34_08705 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 8.26RR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 8.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.32GRR39 pKa = 11.84ARR41 pKa = 11.84LTPP44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 8.26RR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 8.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.32GRR39 pKa = 11.84ARR41 pKa = 11.84LTPP44 pKa = 3.93
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1191587 |
33 |
2193 |
319.3 |
34.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.168 ± 0.065 | 0.91 ± 0.011 |
5.536 ± 0.034 | 6.798 ± 0.045 |
3.129 ± 0.028 | 9.074 ± 0.042 |
2.263 ± 0.019 | 3.899 ± 0.031 |
1.563 ± 0.025 | 11.325 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.031 ± 0.019 | 1.94 ± 0.021 |
5.548 ± 0.03 | 3.148 ± 0.023 |
9.16 ± 0.045 | 4.543 ± 0.028 |
4.727 ± 0.02 | 7.602 ± 0.03 |
1.437 ± 0.018 | 2.2 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
