
Streptomyces hoynatensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
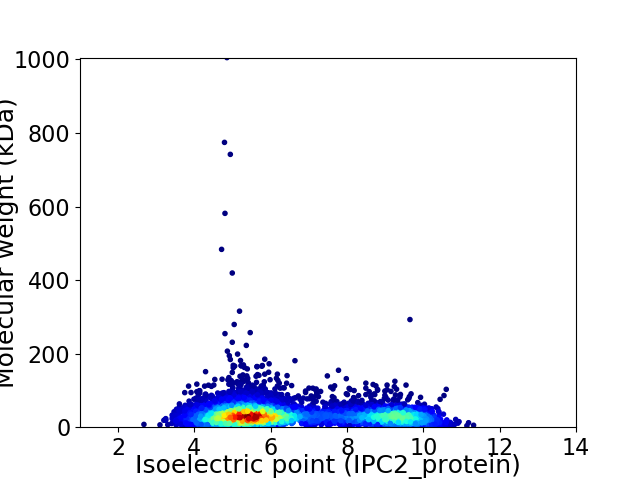
Virtual 2D-PAGE plot for 5959 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A9Z7F1|A0A3A9Z7F1_9ACTN Thiamine pyrophosphate-dependent dehydrogenase E1 component subunit alpha OS=Streptomyces hoynatensis OX=1141874 GN=D7294_11780 PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84KK3 pKa = 8.71PVLALCTATALLAGAVGCSSDD24 pKa = 4.73DD25 pKa = 4.16RR26 pKa = 11.84PDD28 pKa = 3.02QRR30 pKa = 11.84APSEE34 pKa = 4.37SPSGSEE40 pKa = 3.44QTPPPEE46 pKa = 5.69APDD49 pKa = 3.85APVLEE54 pKa = 4.79ALPEE58 pKa = 4.43EE59 pKa = 4.94IPAEE63 pKa = 3.99LDD65 pKa = 3.27PYY67 pKa = 11.28YY68 pKa = 10.16EE69 pKa = 4.45QQLSWQPCEE78 pKa = 5.0AGFEE82 pKa = 4.4CATMRR87 pKa = 11.84VPLDD91 pKa = 3.77YY92 pKa = 11.47EE93 pKa = 4.51NVDD96 pKa = 3.42PDD98 pKa = 3.4QDD100 pKa = 4.07LEE102 pKa = 4.44LTVTRR107 pKa = 11.84SQATDD112 pKa = 3.42PSSRR116 pKa = 11.84IGSLLVNPGGPGASAVDD133 pKa = 4.36FAQDD137 pKa = 3.0SASYY141 pKa = 9.98IFSSEE146 pKa = 4.05VTSHH150 pKa = 6.3YY151 pKa = 11.14DD152 pKa = 3.28VVAVDD157 pKa = 3.74PRR159 pKa = 11.84GTGASEE165 pKa = 3.97PVQCMSGEE173 pKa = 4.09QMDD176 pKa = 4.63EE177 pKa = 3.8FTLNDD182 pKa = 4.11RR183 pKa = 11.84TPDD186 pKa = 3.55SQDD189 pKa = 3.65EE190 pKa = 4.25IDD192 pKa = 3.94TLLSSFDD199 pKa = 3.57EE200 pKa = 4.92FAASCAEE207 pKa = 3.89HH208 pKa = 7.26SGDD211 pKa = 4.04LLGHH215 pKa = 6.85ISTIEE220 pKa = 3.97SARR223 pKa = 11.84DD224 pKa = 3.4MDD226 pKa = 4.31VLRR229 pKa = 11.84ALLGDD234 pKa = 4.09DD235 pKa = 3.17QLHH238 pKa = 5.41YY239 pKa = 11.35VGFSYY244 pKa = 8.91GTKK247 pKa = 10.3LGAVYY252 pKa = 10.54AGLFPQRR259 pKa = 11.84VGRR262 pKa = 11.84LVLDD266 pKa = 4.06GAIDD270 pKa = 3.77PRR272 pKa = 11.84FPTLDD277 pKa = 3.43TDD279 pKa = 3.94RR280 pKa = 11.84QQAGGFEE287 pKa = 4.04TAFRR291 pKa = 11.84SFAEE295 pKa = 4.62DD296 pKa = 3.36CASRR300 pKa = 11.84PDD302 pKa = 4.34CPLGTQGGDD311 pKa = 3.39DD312 pKa = 3.41ASQRR316 pKa = 11.84LLDD319 pKa = 4.11FFEE322 pKa = 4.5QVDD325 pKa = 4.26AEE327 pKa = 4.38PLPTSDD333 pKa = 4.02PDD335 pKa = 3.57RR336 pKa = 11.84PLTEE340 pKa = 4.42SLASTGVSMAMYY352 pKa = 10.15SDD354 pKa = 4.03YY355 pKa = 10.36MWEE358 pKa = 4.0QLRR361 pKa = 11.84EE362 pKa = 4.11ALRR365 pKa = 11.84AAISQGNGDD374 pKa = 4.13QLLALADD381 pKa = 4.27EE382 pKa = 4.76YY383 pKa = 11.66NSRR386 pKa = 11.84EE387 pKa = 3.77PDD389 pKa = 3.22GSYY392 pKa = 9.57GTDD395 pKa = 2.68MFAFPAISCLDD406 pKa = 3.71SPAGNATEE414 pKa = 5.46DD415 pKa = 3.75EE416 pKa = 4.51VEE418 pKa = 4.39EE419 pKa = 3.91NLPAYY424 pKa = 9.82EE425 pKa = 4.1EE426 pKa = 4.32ASPTFGRR433 pKa = 11.84DD434 pKa = 3.36FAWATLQCAAWPVEE448 pKa = 4.11PTGAPVTIEE457 pKa = 4.12AEE459 pKa = 4.17GANDD463 pKa = 3.14IVVVGTTRR471 pKa = 11.84DD472 pKa = 3.27PATPYY477 pKa = 10.63SWAQGLADD485 pKa = 3.72QLSSGVLLTYY495 pKa = 10.84DD496 pKa = 3.89GDD498 pKa = 3.53GHH500 pKa = 5.35TAYY503 pKa = 10.24GGTSPCIDD511 pKa = 3.52AAVNAYY517 pKa = 10.02LLEE520 pKa = 4.47GQTPEE525 pKa = 4.85EE526 pKa = 4.6GTTCC530 pKa = 4.53
MM1 pKa = 7.9RR2 pKa = 11.84KK3 pKa = 8.71PVLALCTATALLAGAVGCSSDD24 pKa = 4.73DD25 pKa = 4.16RR26 pKa = 11.84PDD28 pKa = 3.02QRR30 pKa = 11.84APSEE34 pKa = 4.37SPSGSEE40 pKa = 3.44QTPPPEE46 pKa = 5.69APDD49 pKa = 3.85APVLEE54 pKa = 4.79ALPEE58 pKa = 4.43EE59 pKa = 4.94IPAEE63 pKa = 3.99LDD65 pKa = 3.27PYY67 pKa = 11.28YY68 pKa = 10.16EE69 pKa = 4.45QQLSWQPCEE78 pKa = 5.0AGFEE82 pKa = 4.4CATMRR87 pKa = 11.84VPLDD91 pKa = 3.77YY92 pKa = 11.47EE93 pKa = 4.51NVDD96 pKa = 3.42PDD98 pKa = 3.4QDD100 pKa = 4.07LEE102 pKa = 4.44LTVTRR107 pKa = 11.84SQATDD112 pKa = 3.42PSSRR116 pKa = 11.84IGSLLVNPGGPGASAVDD133 pKa = 4.36FAQDD137 pKa = 3.0SASYY141 pKa = 9.98IFSSEE146 pKa = 4.05VTSHH150 pKa = 6.3YY151 pKa = 11.14DD152 pKa = 3.28VVAVDD157 pKa = 3.74PRR159 pKa = 11.84GTGASEE165 pKa = 3.97PVQCMSGEE173 pKa = 4.09QMDD176 pKa = 4.63EE177 pKa = 3.8FTLNDD182 pKa = 4.11RR183 pKa = 11.84TPDD186 pKa = 3.55SQDD189 pKa = 3.65EE190 pKa = 4.25IDD192 pKa = 3.94TLLSSFDD199 pKa = 3.57EE200 pKa = 4.92FAASCAEE207 pKa = 3.89HH208 pKa = 7.26SGDD211 pKa = 4.04LLGHH215 pKa = 6.85ISTIEE220 pKa = 3.97SARR223 pKa = 11.84DD224 pKa = 3.4MDD226 pKa = 4.31VLRR229 pKa = 11.84ALLGDD234 pKa = 4.09DD235 pKa = 3.17QLHH238 pKa = 5.41YY239 pKa = 11.35VGFSYY244 pKa = 8.91GTKK247 pKa = 10.3LGAVYY252 pKa = 10.54AGLFPQRR259 pKa = 11.84VGRR262 pKa = 11.84LVLDD266 pKa = 4.06GAIDD270 pKa = 3.77PRR272 pKa = 11.84FPTLDD277 pKa = 3.43TDD279 pKa = 3.94RR280 pKa = 11.84QQAGGFEE287 pKa = 4.04TAFRR291 pKa = 11.84SFAEE295 pKa = 4.62DD296 pKa = 3.36CASRR300 pKa = 11.84PDD302 pKa = 4.34CPLGTQGGDD311 pKa = 3.39DD312 pKa = 3.41ASQRR316 pKa = 11.84LLDD319 pKa = 4.11FFEE322 pKa = 4.5QVDD325 pKa = 4.26AEE327 pKa = 4.38PLPTSDD333 pKa = 4.02PDD335 pKa = 3.57RR336 pKa = 11.84PLTEE340 pKa = 4.42SLASTGVSMAMYY352 pKa = 10.15SDD354 pKa = 4.03YY355 pKa = 10.36MWEE358 pKa = 4.0QLRR361 pKa = 11.84EE362 pKa = 4.11ALRR365 pKa = 11.84AAISQGNGDD374 pKa = 4.13QLLALADD381 pKa = 4.27EE382 pKa = 4.76YY383 pKa = 11.66NSRR386 pKa = 11.84EE387 pKa = 3.77PDD389 pKa = 3.22GSYY392 pKa = 9.57GTDD395 pKa = 2.68MFAFPAISCLDD406 pKa = 3.71SPAGNATEE414 pKa = 5.46DD415 pKa = 3.75EE416 pKa = 4.51VEE418 pKa = 4.39EE419 pKa = 3.91NLPAYY424 pKa = 9.82EE425 pKa = 4.1EE426 pKa = 4.32ASPTFGRR433 pKa = 11.84DD434 pKa = 3.36FAWATLQCAAWPVEE448 pKa = 4.11PTGAPVTIEE457 pKa = 4.12AEE459 pKa = 4.17GANDD463 pKa = 3.14IVVVGTTRR471 pKa = 11.84DD472 pKa = 3.27PATPYY477 pKa = 10.63SWAQGLADD485 pKa = 3.72QLSSGVLLTYY495 pKa = 10.84DD496 pKa = 3.89GDD498 pKa = 3.53GHH500 pKa = 5.35TAYY503 pKa = 10.24GGTSPCIDD511 pKa = 3.52AAVNAYY517 pKa = 10.02LLEE520 pKa = 4.47GQTPEE525 pKa = 4.85EE526 pKa = 4.6GTTCC530 pKa = 4.53
Molecular weight: 56.53 kDa
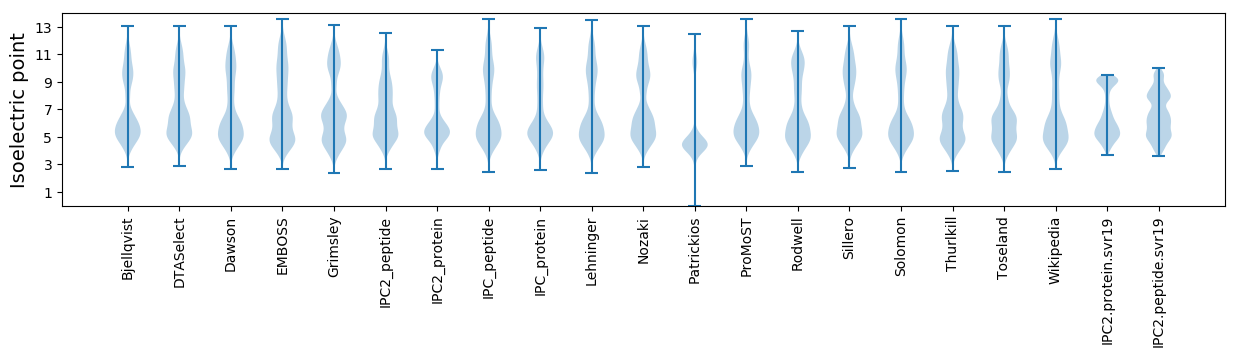
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A9YNW6|A0A3A9YNW6_9ACTN Polyketide cyclase OS=Streptomyces hoynatensis OX=1141874 GN=D7294_29040 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.82GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.82GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2017824 |
18 |
9676 |
338.6 |
36.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.894 ± 0.057 | 0.797 ± 0.009 |
5.406 ± 0.028 | 6.259 ± 0.032 |
2.648 ± 0.019 | 10.163 ± 0.032 |
2.258 ± 0.014 | 2.736 ± 0.024 |
1.33 ± 0.018 | 10.856 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.547 ± 0.013 | 1.56 ± 0.016 |
6.608 ± 0.037 | 2.606 ± 0.02 |
8.912 ± 0.042 | 4.604 ± 0.023 |
5.463 ± 0.026 | 7.871 ± 0.032 |
1.529 ± 0.012 | 1.954 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
