
Niakha virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sripuvirus; Niakha sripuvirus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
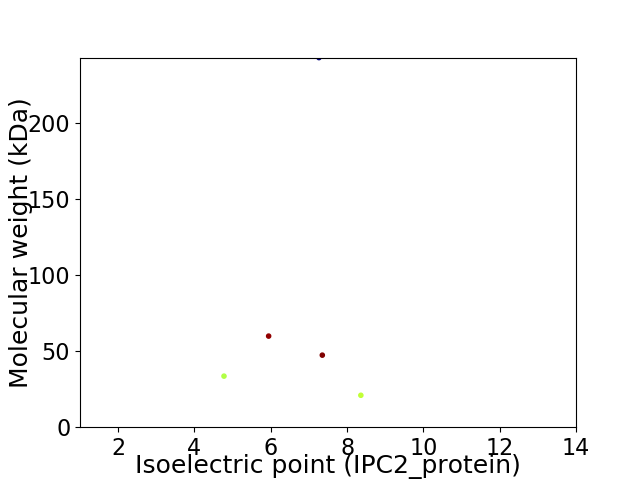
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
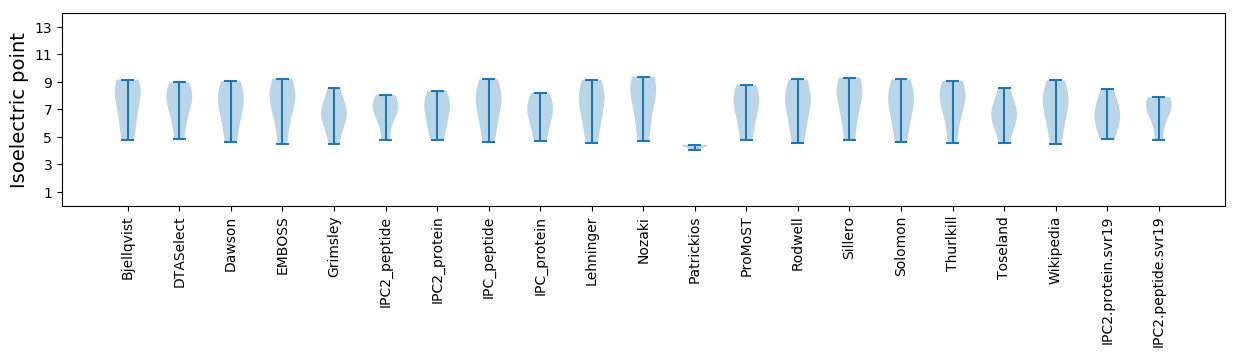
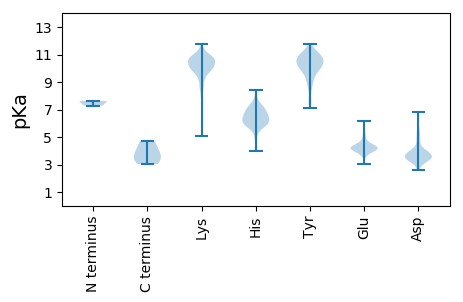
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9ZRL9|R9ZRL9_9RHAB M OS=Niakha virus OX=1348439 PE=4 SV=1
MM1 pKa = 7.37SFDD4 pKa = 3.77AFEE7 pKa = 4.48FPSDD11 pKa = 3.27IAKK14 pKa = 10.27VAIGTAIKK22 pKa = 10.31ADD24 pKa = 3.14IMEE27 pKa = 4.5EE28 pKa = 4.12EE29 pKa = 4.28NDD31 pKa = 3.63VEE33 pKa = 4.54YY34 pKa = 8.53TTDD37 pKa = 3.27AAEE40 pKa = 4.62AKK42 pKa = 8.78TKK44 pKa = 10.47QDD46 pKa = 2.83IQIPEE51 pKa = 4.28SEE53 pKa = 5.03GGWTWEE59 pKa = 4.02NPVEE63 pKa = 4.4SGSKK67 pKa = 10.12GKK69 pKa = 9.79EE70 pKa = 3.99KK71 pKa = 9.77KK72 pKa = 8.72TLTYY76 pKa = 10.86SSDD79 pKa = 3.51EE80 pKa = 4.06EE81 pKa = 4.71SGNEE85 pKa = 3.89TDD87 pKa = 5.67DD88 pKa = 5.29KK89 pKa = 11.33EE90 pKa = 4.25DD91 pKa = 4.06SKK93 pKa = 11.75DD94 pKa = 3.27QTEE97 pKa = 4.12ITEE100 pKa = 4.36SGDD103 pKa = 3.27TFGPLEE109 pKa = 4.05YY110 pKa = 9.91PKK112 pKa = 9.76MVEE115 pKa = 3.44LTYY118 pKa = 10.07PSKK121 pKa = 10.89SFRR124 pKa = 11.84NKK126 pKa = 10.71DD127 pKa = 3.27PGKK130 pKa = 9.81LFEE133 pKa = 4.2WAITSLFDD141 pKa = 3.39QLGVRR146 pKa = 11.84KK147 pKa = 9.27LHH149 pKa = 6.59FKK151 pKa = 9.81TDD153 pKa = 3.62RR154 pKa = 11.84KK155 pKa = 10.7KK156 pKa = 10.1GTCQIMFDD164 pKa = 5.02GINSLKK170 pKa = 10.31FGRR173 pKa = 11.84ALEE176 pKa = 4.19PEE178 pKa = 3.95QLAFTKK184 pKa = 10.41KK185 pKa = 10.24PEE187 pKa = 4.22TPPPHH192 pKa = 7.07SSPKK196 pKa = 9.97KK197 pKa = 8.21PSAPRR202 pKa = 11.84LVEE205 pKa = 4.02VIEE208 pKa = 4.25AQKK211 pKa = 9.07EE212 pKa = 4.17TRR214 pKa = 11.84TVFKK218 pKa = 10.44EE219 pKa = 4.55GPSPQKK225 pKa = 9.95MQFLLEE231 pKa = 4.18LKK233 pKa = 10.38KK234 pKa = 10.74GIRR237 pKa = 11.84VRR239 pKa = 11.84DD240 pKa = 3.4IEE242 pKa = 4.8GDD244 pKa = 3.85MVCVRR249 pKa = 11.84LRR251 pKa = 11.84DD252 pKa = 3.56LAISEE257 pKa = 4.41EE258 pKa = 4.43EE259 pKa = 4.01VLGTEE264 pKa = 5.09FSGDD268 pKa = 3.57MDD270 pKa = 4.02PVTWLSEE277 pKa = 4.11LKK279 pKa = 10.79DD280 pKa = 3.55QFPHH284 pKa = 6.96LKK286 pKa = 10.19RR287 pKa = 11.84ILAMVDD293 pKa = 3.95FEE295 pKa = 4.62EE296 pKa = 4.55PP297 pKa = 3.04
MM1 pKa = 7.37SFDD4 pKa = 3.77AFEE7 pKa = 4.48FPSDD11 pKa = 3.27IAKK14 pKa = 10.27VAIGTAIKK22 pKa = 10.31ADD24 pKa = 3.14IMEE27 pKa = 4.5EE28 pKa = 4.12EE29 pKa = 4.28NDD31 pKa = 3.63VEE33 pKa = 4.54YY34 pKa = 8.53TTDD37 pKa = 3.27AAEE40 pKa = 4.62AKK42 pKa = 8.78TKK44 pKa = 10.47QDD46 pKa = 2.83IQIPEE51 pKa = 4.28SEE53 pKa = 5.03GGWTWEE59 pKa = 4.02NPVEE63 pKa = 4.4SGSKK67 pKa = 10.12GKK69 pKa = 9.79EE70 pKa = 3.99KK71 pKa = 9.77KK72 pKa = 8.72TLTYY76 pKa = 10.86SSDD79 pKa = 3.51EE80 pKa = 4.06EE81 pKa = 4.71SGNEE85 pKa = 3.89TDD87 pKa = 5.67DD88 pKa = 5.29KK89 pKa = 11.33EE90 pKa = 4.25DD91 pKa = 4.06SKK93 pKa = 11.75DD94 pKa = 3.27QTEE97 pKa = 4.12ITEE100 pKa = 4.36SGDD103 pKa = 3.27TFGPLEE109 pKa = 4.05YY110 pKa = 9.91PKK112 pKa = 9.76MVEE115 pKa = 3.44LTYY118 pKa = 10.07PSKK121 pKa = 10.89SFRR124 pKa = 11.84NKK126 pKa = 10.71DD127 pKa = 3.27PGKK130 pKa = 9.81LFEE133 pKa = 4.2WAITSLFDD141 pKa = 3.39QLGVRR146 pKa = 11.84KK147 pKa = 9.27LHH149 pKa = 6.59FKK151 pKa = 9.81TDD153 pKa = 3.62RR154 pKa = 11.84KK155 pKa = 10.7KK156 pKa = 10.1GTCQIMFDD164 pKa = 5.02GINSLKK170 pKa = 10.31FGRR173 pKa = 11.84ALEE176 pKa = 4.19PEE178 pKa = 3.95QLAFTKK184 pKa = 10.41KK185 pKa = 10.24PEE187 pKa = 4.22TPPPHH192 pKa = 7.07SSPKK196 pKa = 9.97KK197 pKa = 8.21PSAPRR202 pKa = 11.84LVEE205 pKa = 4.02VIEE208 pKa = 4.25AQKK211 pKa = 9.07EE212 pKa = 4.17TRR214 pKa = 11.84TVFKK218 pKa = 10.44EE219 pKa = 4.55GPSPQKK225 pKa = 9.95MQFLLEE231 pKa = 4.18LKK233 pKa = 10.38KK234 pKa = 10.74GIRR237 pKa = 11.84VRR239 pKa = 11.84DD240 pKa = 3.4IEE242 pKa = 4.8GDD244 pKa = 3.85MVCVRR249 pKa = 11.84LRR251 pKa = 11.84DD252 pKa = 3.56LAISEE257 pKa = 4.41EE258 pKa = 4.43EE259 pKa = 4.01VLGTEE264 pKa = 5.09FSGDD268 pKa = 3.57MDD270 pKa = 4.02PVTWLSEE277 pKa = 4.11LKK279 pKa = 10.79DD280 pKa = 3.55QFPHH284 pKa = 6.96LKK286 pKa = 10.19RR287 pKa = 11.84ILAMVDD293 pKa = 3.95FEE295 pKa = 4.62EE296 pKa = 4.55PP297 pKa = 3.04
Molecular weight: 33.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9ZRW4|R9ZRW4_9RHAB GDP polyribonucleotidyltransferase OS=Niakha virus OX=1348439 PE=4 SV=1
MM1 pKa = 7.51NFLFKK6 pKa = 10.36KK7 pKa = 9.93KK8 pKa = 10.09KK9 pKa = 9.95SSEE12 pKa = 4.31SLWGNSSKK20 pKa = 11.24ALVSAPIMKK29 pKa = 10.02NYY31 pKa = 9.91KK32 pKa = 9.23VEE34 pKa = 4.49CKK36 pKa = 10.04IEE38 pKa = 4.23LNVHH42 pKa = 5.88NKK44 pKa = 9.32VNGPDD49 pKa = 4.48DD50 pKa = 3.33IVDD53 pKa = 3.96IYY55 pKa = 11.35RR56 pKa = 11.84DD57 pKa = 3.69YY58 pKa = 11.33CSNYY62 pKa = 9.71CGVIQSRR69 pKa = 11.84PLHH72 pKa = 5.87CFIMALGICHH82 pKa = 6.64IQRR85 pKa = 11.84KK86 pKa = 8.73QGMSQGGDD94 pKa = 3.36YY95 pKa = 10.96LCEE98 pKa = 3.98YY99 pKa = 9.44KK100 pKa = 11.0GKK102 pKa = 9.74IQLQLGEE109 pKa = 4.25GADD112 pKa = 3.71PGVCRR117 pKa = 11.84SYY119 pKa = 10.61QIHH122 pKa = 7.21RR123 pKa = 11.84KK124 pKa = 9.44RR125 pKa = 11.84ISHH128 pKa = 6.99PAMLAYY134 pKa = 9.73ILGNTVFEE142 pKa = 4.86EE143 pKa = 4.29ISDD146 pKa = 3.76EE147 pKa = 4.32GVGSVEE153 pKa = 4.1TMTKK157 pKa = 9.67EE158 pKa = 3.92GRR160 pKa = 11.84IEE162 pKa = 4.15FKK164 pKa = 10.8KK165 pKa = 10.63LVAASPFILRR175 pKa = 11.84SGKK178 pKa = 10.2SRR180 pKa = 11.84TQICARR186 pKa = 11.84PP187 pKa = 3.39
MM1 pKa = 7.51NFLFKK6 pKa = 10.36KK7 pKa = 9.93KK8 pKa = 10.09KK9 pKa = 9.95SSEE12 pKa = 4.31SLWGNSSKK20 pKa = 11.24ALVSAPIMKK29 pKa = 10.02NYY31 pKa = 9.91KK32 pKa = 9.23VEE34 pKa = 4.49CKK36 pKa = 10.04IEE38 pKa = 4.23LNVHH42 pKa = 5.88NKK44 pKa = 9.32VNGPDD49 pKa = 4.48DD50 pKa = 3.33IVDD53 pKa = 3.96IYY55 pKa = 11.35RR56 pKa = 11.84DD57 pKa = 3.69YY58 pKa = 11.33CSNYY62 pKa = 9.71CGVIQSRR69 pKa = 11.84PLHH72 pKa = 5.87CFIMALGICHH82 pKa = 6.64IQRR85 pKa = 11.84KK86 pKa = 8.73QGMSQGGDD94 pKa = 3.36YY95 pKa = 10.96LCEE98 pKa = 3.98YY99 pKa = 9.44KK100 pKa = 11.0GKK102 pKa = 9.74IQLQLGEE109 pKa = 4.25GADD112 pKa = 3.71PGVCRR117 pKa = 11.84SYY119 pKa = 10.61QIHH122 pKa = 7.21RR123 pKa = 11.84KK124 pKa = 9.44RR125 pKa = 11.84ISHH128 pKa = 6.99PAMLAYY134 pKa = 9.73ILGNTVFEE142 pKa = 4.86EE143 pKa = 4.29ISDD146 pKa = 3.76EE147 pKa = 4.32GVGSVEE153 pKa = 4.1TMTKK157 pKa = 9.67EE158 pKa = 3.92GRR160 pKa = 11.84IEE162 pKa = 4.15FKK164 pKa = 10.8KK165 pKa = 10.63LVAASPFILRR175 pKa = 11.84SGKK178 pKa = 10.2SRR180 pKa = 11.84TQICARR186 pKa = 11.84PP187 pKa = 3.39
Molecular weight: 21.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3547 |
187 |
2106 |
709.4 |
81.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.891 ± 0.633 | 2.453 ± 0.302 |
5.469 ± 0.361 | 6.879 ± 0.86 |
4.877 ± 0.149 | 5.892 ± 0.853 |
2.058 ± 0.344 | 7.387 ± 0.797 |
7.753 ± 0.568 | 9.191 ± 0.883 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.763 ± 0.083 | 4.793 ± 0.821 |
4.257 ± 0.629 | 3.327 ± 0.314 |
4.624 ± 0.438 | 8.007 ± 0.372 |
5.695 ± 0.442 | 5.639 ± 0.259 |
1.663 ± 0.191 | 3.383 ± 0.374 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
