
Donkey orchid symptomless virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Alphaflexiviridae; Platypuvirus
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
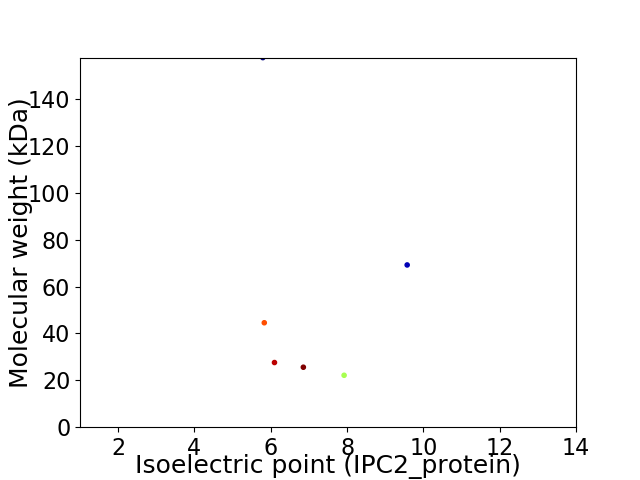
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
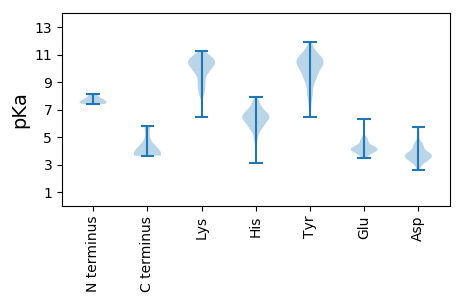
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|V5LWR9|V5LWR9_9VIRU RNA-directed RNA polymerase OS=Donkey orchid symptomless virus OX=1400526 PE=4 SV=1
MM1 pKa = 7.45SLALCRR7 pKa = 11.84QSCHH11 pKa = 4.75SCYY14 pKa = 10.42VFQFFEE20 pKa = 4.04FQMAIINPVTSVRR33 pKa = 11.84WAQMLNNHH41 pKa = 5.45TRR43 pKa = 11.84ASPKK47 pKa = 9.67VAHH50 pKa = 6.2FQNGSDD56 pKa = 3.22CWAPLNFDD64 pKa = 3.06EE65 pKa = 6.33RR66 pKa = 11.84IYY68 pKa = 11.59VDD70 pKa = 4.13LQTGSSFQSSIPVFKK85 pKa = 9.02TTSTDD90 pKa = 3.19NVALLIFAAAYY101 pKa = 9.12DD102 pKa = 3.87RR103 pKa = 11.84RR104 pKa = 11.84ADD106 pKa = 3.93GPFHH110 pKa = 7.57LLGTDD115 pKa = 3.3NNGYY119 pKa = 10.57LYY121 pKa = 9.18TLPSGGTRR129 pKa = 11.84AIKK132 pKa = 10.63VLDD135 pKa = 3.54SFRR138 pKa = 11.84ITVRR142 pKa = 11.84PLEE145 pKa = 4.08QPYY148 pKa = 10.66VDD150 pKa = 3.58AKK152 pKa = 10.39LYY154 pKa = 10.37HH155 pKa = 6.34YY156 pKa = 10.45SVSDD160 pKa = 3.63NFPLSEE166 pKa = 3.93GQARR170 pKa = 11.84IRR172 pKa = 11.84EE173 pKa = 4.14LTIQVEE179 pKa = 4.77TLTSTNEE186 pKa = 3.6DD187 pKa = 3.36LTRR190 pKa = 11.84LNTTYY195 pKa = 10.97SSRR198 pKa = 11.84IDD200 pKa = 3.75SLQKK204 pKa = 11.15DD205 pKa = 3.81NDD207 pKa = 3.76RR208 pKa = 11.84VSAANSTLSDD218 pKa = 3.45EE219 pKa = 4.92KK220 pKa = 10.97SACFSKK226 pKa = 10.61LAQATDD232 pKa = 3.73VIRR235 pKa = 11.84QLRR238 pKa = 11.84EE239 pKa = 3.68TNALALGQIGEE250 pKa = 4.27LSSLNEE256 pKa = 3.9VLGSKK261 pKa = 10.56NDD263 pKa = 4.12DD264 pKa = 3.28LTEE267 pKa = 4.08AQRR270 pKa = 11.84LLEE273 pKa = 4.09TSLAVIEE280 pKa = 4.47TKK282 pKa = 10.64NRR284 pKa = 11.84EE285 pKa = 4.12LTVEE289 pKa = 4.13VAEE292 pKa = 4.79LKK294 pKa = 10.91AKK296 pKa = 10.87LEE298 pKa = 4.22ACKK301 pKa = 8.84PTSISPLPLTHH312 pKa = 7.13PALGVNHH319 pKa = 7.13LYY321 pKa = 10.31CWFRR325 pKa = 11.84HH326 pKa = 6.0AGTQLKK332 pKa = 10.02RR333 pKa = 11.84RR334 pKa = 11.84ARR336 pKa = 11.84TDD338 pKa = 3.33LPTMSNYY345 pKa = 9.42IVASRR350 pKa = 11.84IEE352 pKa = 4.25RR353 pKa = 11.84FAEE356 pKa = 3.79LTLIEE361 pKa = 4.31VALLVDD367 pKa = 3.55NTAIKK372 pKa = 9.81IASVTTTGSIISTPQVDD389 pKa = 4.53SLICLHH395 pKa = 7.1LGICFCC401 pKa = 5.81
MM1 pKa = 7.45SLALCRR7 pKa = 11.84QSCHH11 pKa = 4.75SCYY14 pKa = 10.42VFQFFEE20 pKa = 4.04FQMAIINPVTSVRR33 pKa = 11.84WAQMLNNHH41 pKa = 5.45TRR43 pKa = 11.84ASPKK47 pKa = 9.67VAHH50 pKa = 6.2FQNGSDD56 pKa = 3.22CWAPLNFDD64 pKa = 3.06EE65 pKa = 6.33RR66 pKa = 11.84IYY68 pKa = 11.59VDD70 pKa = 4.13LQTGSSFQSSIPVFKK85 pKa = 9.02TTSTDD90 pKa = 3.19NVALLIFAAAYY101 pKa = 9.12DD102 pKa = 3.87RR103 pKa = 11.84RR104 pKa = 11.84ADD106 pKa = 3.93GPFHH110 pKa = 7.57LLGTDD115 pKa = 3.3NNGYY119 pKa = 10.57LYY121 pKa = 9.18TLPSGGTRR129 pKa = 11.84AIKK132 pKa = 10.63VLDD135 pKa = 3.54SFRR138 pKa = 11.84ITVRR142 pKa = 11.84PLEE145 pKa = 4.08QPYY148 pKa = 10.66VDD150 pKa = 3.58AKK152 pKa = 10.39LYY154 pKa = 10.37HH155 pKa = 6.34YY156 pKa = 10.45SVSDD160 pKa = 3.63NFPLSEE166 pKa = 3.93GQARR170 pKa = 11.84IRR172 pKa = 11.84EE173 pKa = 4.14LTIQVEE179 pKa = 4.77TLTSTNEE186 pKa = 3.6DD187 pKa = 3.36LTRR190 pKa = 11.84LNTTYY195 pKa = 10.97SSRR198 pKa = 11.84IDD200 pKa = 3.75SLQKK204 pKa = 11.15DD205 pKa = 3.81NDD207 pKa = 3.76RR208 pKa = 11.84VSAANSTLSDD218 pKa = 3.45EE219 pKa = 4.92KK220 pKa = 10.97SACFSKK226 pKa = 10.61LAQATDD232 pKa = 3.73VIRR235 pKa = 11.84QLRR238 pKa = 11.84EE239 pKa = 3.68TNALALGQIGEE250 pKa = 4.27LSSLNEE256 pKa = 3.9VLGSKK261 pKa = 10.56NDD263 pKa = 4.12DD264 pKa = 3.28LTEE267 pKa = 4.08AQRR270 pKa = 11.84LLEE273 pKa = 4.09TSLAVIEE280 pKa = 4.47TKK282 pKa = 10.64NRR284 pKa = 11.84EE285 pKa = 4.12LTVEE289 pKa = 4.13VAEE292 pKa = 4.79LKK294 pKa = 10.91AKK296 pKa = 10.87LEE298 pKa = 4.22ACKK301 pKa = 8.84PTSISPLPLTHH312 pKa = 7.13PALGVNHH319 pKa = 7.13LYY321 pKa = 10.31CWFRR325 pKa = 11.84HH326 pKa = 6.0AGTQLKK332 pKa = 10.02RR333 pKa = 11.84RR334 pKa = 11.84ARR336 pKa = 11.84TDD338 pKa = 3.33LPTMSNYY345 pKa = 9.42IVASRR350 pKa = 11.84IEE352 pKa = 4.25RR353 pKa = 11.84FAEE356 pKa = 3.79LTLIEE361 pKa = 4.31VALLVDD367 pKa = 3.55NTAIKK372 pKa = 9.81IASVTTTGSIISTPQVDD389 pKa = 4.53SLICLHH395 pKa = 7.1LGICFCC401 pKa = 5.81
Molecular weight: 44.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V5LUG3|V5LUG3_9VIRU Movement protein OS=Donkey orchid symptomless virus OX=1400526 PE=4 SV=1
MM1 pKa = 8.1RR2 pKa = 11.84EE3 pKa = 3.95NQPNSSITAANEE15 pKa = 3.77TRR17 pKa = 11.84SPLEE21 pKa = 4.03LEE23 pKa = 3.98KK24 pKa = 11.04HH25 pKa = 5.6RR26 pKa = 11.84VDD28 pKa = 4.8LQSSDD33 pKa = 2.58HH34 pKa = 6.71RR35 pKa = 11.84LRR37 pKa = 11.84MSCYY41 pKa = 10.09LKK43 pKa = 9.65STQPSRR49 pKa = 11.84MSTSRR54 pKa = 11.84QSSRR58 pKa = 11.84LGTGPNTTMRR68 pKa = 11.84STKK71 pKa = 9.73PGRR74 pKa = 11.84PTHH77 pKa = 5.47TTSTMRR83 pKa = 11.84PPNFSLGSGFPPFPAMRR100 pKa = 11.84PPSSTAMEE108 pKa = 4.01KK109 pKa = 9.41TRR111 pKa = 11.84PSKK114 pKa = 9.74ITSLPTSNTNSQAPAASASSSPLSCSSLATTPEE147 pKa = 3.97PRR149 pKa = 11.84TSSSTLTLLRR159 pKa = 11.84RR160 pKa = 11.84TSTDD164 pKa = 2.63SPRR167 pKa = 11.84TTLSPTSSNSTTPPPSCRR185 pKa = 11.84MLCTTTPQASSSTSSKK201 pKa = 8.28EE202 pKa = 3.88TLVFGSFTPQLLSQLKK218 pKa = 9.68SCTSTLPFTLAYY230 pKa = 8.17TPSSITTTTSSHH242 pKa = 6.19TSQSPPPPEE251 pKa = 4.35PTPKK255 pKa = 10.56ALPALTGSNTPTSPGAKK272 pKa = 8.59PKK274 pKa = 10.77CPALSSKK281 pKa = 10.49PLGPTMSSISCAGSFCHH298 pKa = 7.43KK299 pKa = 10.78SGDD302 pKa = 3.9CSNILHH308 pKa = 6.52SSSCPRR314 pKa = 11.84YY315 pKa = 7.08TAWGASIRR323 pKa = 11.84TNPSQKK329 pKa = 10.0RR330 pKa = 11.84SCKK333 pKa = 10.07CFSCMRR339 pKa = 11.84TPSKK343 pKa = 10.68KK344 pKa = 10.64LEE346 pKa = 4.21TSTSGPSSASKK357 pKa = 10.22FRR359 pKa = 11.84RR360 pKa = 11.84TPSTTMTSEE369 pKa = 3.79ISHH372 pKa = 6.36SSRR375 pKa = 11.84TTSSSPPNSVGTQMRR390 pKa = 11.84SPSPTRR396 pKa = 11.84ASSGAYY402 pKa = 8.99QRR404 pKa = 11.84TPRR407 pKa = 11.84TPYY410 pKa = 9.79GAHH413 pKa = 6.36SPHH416 pKa = 7.14SSVQTASKK424 pKa = 8.28NTSVNSSSSRR434 pKa = 11.84SITPSRR440 pKa = 11.84PNATSQTTYY449 pKa = 11.38QLLCTLGSKK458 pKa = 8.43TATQLISPATNCSSLRR474 pKa = 11.84RR475 pKa = 11.84ATAKK479 pKa = 6.47WTRR482 pKa = 11.84RR483 pKa = 11.84PSADD487 pKa = 3.1ANQMYY492 pKa = 10.31RR493 pKa = 11.84IVEE496 pKa = 3.92QTEE499 pKa = 3.86WEE501 pKa = 4.58TRR503 pKa = 11.84CLHH506 pKa = 5.79IMEE509 pKa = 4.95WSFPLQSYY517 pKa = 8.07TMACSLSCPVNATGAIRR534 pKa = 11.84NWRR537 pKa = 11.84VCSNSRR543 pKa = 11.84LSKK546 pKa = 9.21NQAVRR551 pKa = 11.84IMRR554 pKa = 11.84EE555 pKa = 3.59HH556 pKa = 6.54QIFKK560 pKa = 8.91TAHH563 pKa = 6.17PPSTPVSSQSMTHH576 pKa = 5.28IHH578 pKa = 7.58ASRR581 pKa = 11.84QPQPSTSLSASMGHH595 pKa = 5.7RR596 pKa = 11.84HH597 pKa = 6.02SPPTALVVPPTSPLISTEE615 pKa = 3.78AGLALSTDD623 pKa = 3.83GKK625 pKa = 8.94ITPASSSGNTCVMGSEE641 pKa = 4.4TKK643 pKa = 8.84PTATLTTT650 pKa = 3.93
MM1 pKa = 8.1RR2 pKa = 11.84EE3 pKa = 3.95NQPNSSITAANEE15 pKa = 3.77TRR17 pKa = 11.84SPLEE21 pKa = 4.03LEE23 pKa = 3.98KK24 pKa = 11.04HH25 pKa = 5.6RR26 pKa = 11.84VDD28 pKa = 4.8LQSSDD33 pKa = 2.58HH34 pKa = 6.71RR35 pKa = 11.84LRR37 pKa = 11.84MSCYY41 pKa = 10.09LKK43 pKa = 9.65STQPSRR49 pKa = 11.84MSTSRR54 pKa = 11.84QSSRR58 pKa = 11.84LGTGPNTTMRR68 pKa = 11.84STKK71 pKa = 9.73PGRR74 pKa = 11.84PTHH77 pKa = 5.47TTSTMRR83 pKa = 11.84PPNFSLGSGFPPFPAMRR100 pKa = 11.84PPSSTAMEE108 pKa = 4.01KK109 pKa = 9.41TRR111 pKa = 11.84PSKK114 pKa = 9.74ITSLPTSNTNSQAPAASASSSPLSCSSLATTPEE147 pKa = 3.97PRR149 pKa = 11.84TSSSTLTLLRR159 pKa = 11.84RR160 pKa = 11.84TSTDD164 pKa = 2.63SPRR167 pKa = 11.84TTLSPTSSNSTTPPPSCRR185 pKa = 11.84MLCTTTPQASSSTSSKK201 pKa = 8.28EE202 pKa = 3.88TLVFGSFTPQLLSQLKK218 pKa = 9.68SCTSTLPFTLAYY230 pKa = 8.17TPSSITTTTSSHH242 pKa = 6.19TSQSPPPPEE251 pKa = 4.35PTPKK255 pKa = 10.56ALPALTGSNTPTSPGAKK272 pKa = 8.59PKK274 pKa = 10.77CPALSSKK281 pKa = 10.49PLGPTMSSISCAGSFCHH298 pKa = 7.43KK299 pKa = 10.78SGDD302 pKa = 3.9CSNILHH308 pKa = 6.52SSSCPRR314 pKa = 11.84YY315 pKa = 7.08TAWGASIRR323 pKa = 11.84TNPSQKK329 pKa = 10.0RR330 pKa = 11.84SCKK333 pKa = 10.07CFSCMRR339 pKa = 11.84TPSKK343 pKa = 10.68KK344 pKa = 10.64LEE346 pKa = 4.21TSTSGPSSASKK357 pKa = 10.22FRR359 pKa = 11.84RR360 pKa = 11.84TPSTTMTSEE369 pKa = 3.79ISHH372 pKa = 6.36SSRR375 pKa = 11.84TTSSSPPNSVGTQMRR390 pKa = 11.84SPSPTRR396 pKa = 11.84ASSGAYY402 pKa = 8.99QRR404 pKa = 11.84TPRR407 pKa = 11.84TPYY410 pKa = 9.79GAHH413 pKa = 6.36SPHH416 pKa = 7.14SSVQTASKK424 pKa = 8.28NTSVNSSSSRR434 pKa = 11.84SITPSRR440 pKa = 11.84PNATSQTTYY449 pKa = 11.38QLLCTLGSKK458 pKa = 8.43TATQLISPATNCSSLRR474 pKa = 11.84RR475 pKa = 11.84ATAKK479 pKa = 6.47WTRR482 pKa = 11.84RR483 pKa = 11.84PSADD487 pKa = 3.1ANQMYY492 pKa = 10.31RR493 pKa = 11.84IVEE496 pKa = 3.92QTEE499 pKa = 3.86WEE501 pKa = 4.58TRR503 pKa = 11.84CLHH506 pKa = 5.79IMEE509 pKa = 4.95WSFPLQSYY517 pKa = 8.07TMACSLSCPVNATGAIRR534 pKa = 11.84NWRR537 pKa = 11.84VCSNSRR543 pKa = 11.84LSKK546 pKa = 9.21NQAVRR551 pKa = 11.84IMRR554 pKa = 11.84EE555 pKa = 3.59HH556 pKa = 6.54QIFKK560 pKa = 8.91TAHH563 pKa = 6.17PPSTPVSSQSMTHH576 pKa = 5.28IHH578 pKa = 7.58ASRR581 pKa = 11.84QPQPSTSLSASMGHH595 pKa = 5.7RR596 pKa = 11.84HH597 pKa = 6.02SPPTALVVPPTSPLISTEE615 pKa = 3.78AGLALSTDD623 pKa = 3.83GKK625 pKa = 8.94ITPASSSGNTCVMGSEE641 pKa = 4.4TKK643 pKa = 8.84PTATLTTT650 pKa = 3.93
Molecular weight: 69.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3120 |
202 |
1396 |
520.0 |
57.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.365 ± 0.585 | 2.244 ± 0.279 |
4.647 ± 1.051 | 5.321 ± 0.865 |
3.365 ± 0.45 | 4.423 ± 0.307 |
3.814 ± 0.657 | 4.968 ± 0.605 |
4.038 ± 0.166 | 9.135 ± 1.009 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.538 ± 0.437 | 4.103 ± 0.284 |
7.308 ± 0.991 | 3.91 ± 0.087 |
6.378 ± 0.138 | 9.327 ± 2.989 |
8.205 ± 1.744 | 4.872 ± 0.767 |
0.769 ± 0.062 | 3.269 ± 0.6 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
