
Gloeocapsa sp. PCC 7428
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Chroococcales; Chroococcaceae; Gloeocapsa; unclassified Gloeocapsa
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
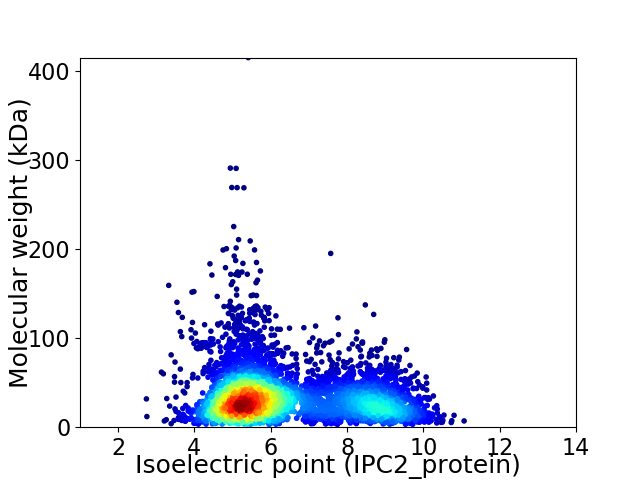
Virtual 2D-PAGE plot for 5003 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
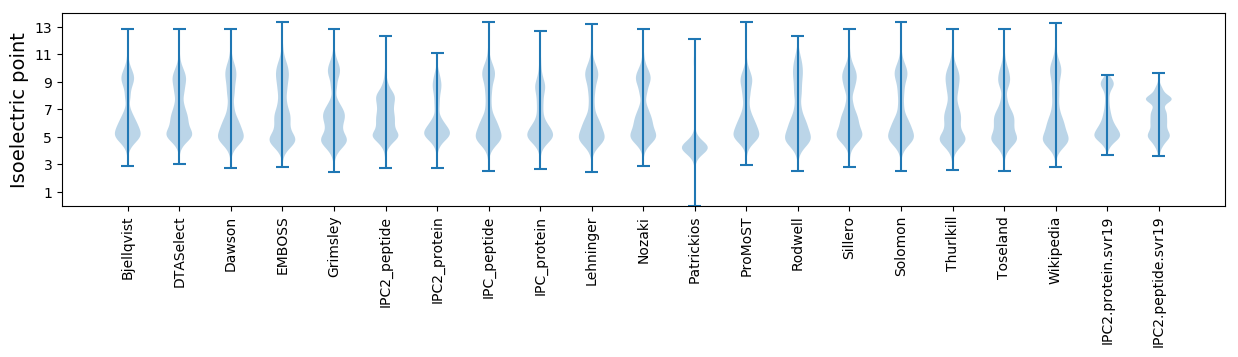
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9XAJ2|K9XAJ2_9CHRO K+transporting ATPase F subunit OS=Gloeocapsa sp. PCC 7428 OX=1173026 GN=Glo7428_0029 PE=4 SV=1
MM1 pKa = 7.7AIIKK5 pKa = 8.02GTAGNDD11 pKa = 3.35NLKK14 pKa = 10.88GGSNNDD20 pKa = 2.97KK21 pKa = 10.71LYY23 pKa = 10.72GYY25 pKa = 10.06EE26 pKa = 5.79GEE28 pKa = 4.29DD29 pKa = 3.65TLNGGAGIDD38 pKa = 3.71TLIGGTGSDD47 pKa = 3.93VYY49 pKa = 10.98IVDD52 pKa = 3.54TTTDD56 pKa = 3.37VIIEE60 pKa = 4.04YY61 pKa = 10.37SGGGAHH67 pKa = 6.7DD68 pKa = 4.04TVKK71 pKa = 10.94SSVSYY76 pKa = 9.75TLGAYY81 pKa = 10.43VNDD84 pKa = 3.82LVLTGSSNINGTGNEE99 pKa = 4.37LDD101 pKa = 4.52NIITGNSGRR110 pKa = 11.84NVLRR114 pKa = 11.84GLAGNDD120 pKa = 3.28QLNGSNDD127 pKa = 3.11KK128 pKa = 11.09RR129 pKa = 11.84SDD131 pKa = 3.36TLYY134 pKa = 11.11GGVGNDD140 pKa = 3.4SLKK143 pKa = 11.15GGSKK147 pKa = 10.61DD148 pKa = 3.4KK149 pKa = 11.19LYY151 pKa = 11.32GGAGNDD157 pKa = 3.15RR158 pKa = 11.84LVGSHH163 pKa = 6.66VGFFDD168 pKa = 4.47NPDD171 pKa = 3.16DD172 pKa = 4.39TIYY175 pKa = 10.5MDD177 pKa = 4.65GGDD180 pKa = 4.08GNDD183 pKa = 3.52YY184 pKa = 10.99LYY186 pKa = 11.04ASHH189 pKa = 7.43DD190 pKa = 3.38AEE192 pKa = 4.64MYY194 pKa = 10.3GGRR197 pKa = 11.84GSDD200 pKa = 3.74TITGGTSVYY209 pKa = 10.27IDD211 pKa = 4.3GGDD214 pKa = 4.14GDD216 pKa = 5.46DD217 pKa = 5.68LLDD220 pKa = 4.27GDD222 pKa = 4.66EE223 pKa = 4.12STIYY227 pKa = 10.34GGKK230 pKa = 10.72GNDD233 pKa = 4.2TITGSYY239 pKa = 9.46SRR241 pKa = 11.84LYY243 pKa = 10.83GGDD246 pKa = 3.45GDD248 pKa = 5.45DD249 pKa = 3.92VLSGIAGMEE258 pKa = 4.07GGQGNDD264 pKa = 3.29TLIGGDD270 pKa = 3.46WDD272 pKa = 3.67SGFIFSKK279 pKa = 8.97PTDD282 pKa = 4.72GIDD285 pKa = 3.45TITNFAADD293 pKa = 4.9RR294 pKa = 11.84YY295 pKa = 9.03HH296 pKa = 7.13KK297 pKa = 10.15IYY299 pKa = 10.91VSASGFGGGLVEE311 pKa = 5.75GEE313 pKa = 4.61LLPGQLQFGTSATEE327 pKa = 3.6ADD329 pKa = 3.94DD330 pKa = 4.42RR331 pKa = 11.84FIYY334 pKa = 8.91DD335 pKa = 3.5TSSGALYY342 pKa = 10.51FDD344 pKa = 3.52VDD346 pKa = 3.99GVLGTGSQVQLAILVGVPDD365 pKa = 4.29LTSSSIYY372 pKa = 8.79VTSS375 pKa = 3.41
MM1 pKa = 7.7AIIKK5 pKa = 8.02GTAGNDD11 pKa = 3.35NLKK14 pKa = 10.88GGSNNDD20 pKa = 2.97KK21 pKa = 10.71LYY23 pKa = 10.72GYY25 pKa = 10.06EE26 pKa = 5.79GEE28 pKa = 4.29DD29 pKa = 3.65TLNGGAGIDD38 pKa = 3.71TLIGGTGSDD47 pKa = 3.93VYY49 pKa = 10.98IVDD52 pKa = 3.54TTTDD56 pKa = 3.37VIIEE60 pKa = 4.04YY61 pKa = 10.37SGGGAHH67 pKa = 6.7DD68 pKa = 4.04TVKK71 pKa = 10.94SSVSYY76 pKa = 9.75TLGAYY81 pKa = 10.43VNDD84 pKa = 3.82LVLTGSSNINGTGNEE99 pKa = 4.37LDD101 pKa = 4.52NIITGNSGRR110 pKa = 11.84NVLRR114 pKa = 11.84GLAGNDD120 pKa = 3.28QLNGSNDD127 pKa = 3.11KK128 pKa = 11.09RR129 pKa = 11.84SDD131 pKa = 3.36TLYY134 pKa = 11.11GGVGNDD140 pKa = 3.4SLKK143 pKa = 11.15GGSKK147 pKa = 10.61DD148 pKa = 3.4KK149 pKa = 11.19LYY151 pKa = 11.32GGAGNDD157 pKa = 3.15RR158 pKa = 11.84LVGSHH163 pKa = 6.66VGFFDD168 pKa = 4.47NPDD171 pKa = 3.16DD172 pKa = 4.39TIYY175 pKa = 10.5MDD177 pKa = 4.65GGDD180 pKa = 4.08GNDD183 pKa = 3.52YY184 pKa = 10.99LYY186 pKa = 11.04ASHH189 pKa = 7.43DD190 pKa = 3.38AEE192 pKa = 4.64MYY194 pKa = 10.3GGRR197 pKa = 11.84GSDD200 pKa = 3.74TITGGTSVYY209 pKa = 10.27IDD211 pKa = 4.3GGDD214 pKa = 4.14GDD216 pKa = 5.46DD217 pKa = 5.68LLDD220 pKa = 4.27GDD222 pKa = 4.66EE223 pKa = 4.12STIYY227 pKa = 10.34GGKK230 pKa = 10.72GNDD233 pKa = 4.2TITGSYY239 pKa = 9.46SRR241 pKa = 11.84LYY243 pKa = 10.83GGDD246 pKa = 3.45GDD248 pKa = 5.45DD249 pKa = 3.92VLSGIAGMEE258 pKa = 4.07GGQGNDD264 pKa = 3.29TLIGGDD270 pKa = 3.46WDD272 pKa = 3.67SGFIFSKK279 pKa = 8.97PTDD282 pKa = 4.72GIDD285 pKa = 3.45TITNFAADD293 pKa = 4.9RR294 pKa = 11.84YY295 pKa = 9.03HH296 pKa = 7.13KK297 pKa = 10.15IYY299 pKa = 10.91VSASGFGGGLVEE311 pKa = 5.75GEE313 pKa = 4.61LLPGQLQFGTSATEE327 pKa = 3.6ADD329 pKa = 3.94DD330 pKa = 4.42RR331 pKa = 11.84FIYY334 pKa = 8.91DD335 pKa = 3.5TSSGALYY342 pKa = 10.51FDD344 pKa = 3.52VDD346 pKa = 3.99GVLGTGSQVQLAILVGVPDD365 pKa = 4.29LTSSSIYY372 pKa = 8.79VTSS375 pKa = 3.41
Molecular weight: 38.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9XCX1|K9XCX1_9CHRO HI0933 family protein OS=Gloeocapsa sp. PCC 7428 OX=1173026 GN=Glo7428_1367 PE=4 SV=1
MM1 pKa = 7.35SRR3 pKa = 11.84TRR5 pKa = 11.84RR6 pKa = 11.84HH7 pKa = 5.26RR8 pKa = 11.84NIRR11 pKa = 11.84AAQLFVIFAGITILVWVLRR30 pKa = 11.84GLGILTFLPGGILWILILLSVITGIWSRR58 pKa = 11.84FQRR61 pKa = 3.78
MM1 pKa = 7.35SRR3 pKa = 11.84TRR5 pKa = 11.84RR6 pKa = 11.84HH7 pKa = 5.26RR8 pKa = 11.84NIRR11 pKa = 11.84AAQLFVIFAGITILVWVLRR30 pKa = 11.84GLGILTFLPGGILWILILLSVITGIWSRR58 pKa = 11.84FQRR61 pKa = 3.78
Molecular weight: 7.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1614380 |
29 |
3699 |
322.7 |
35.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.982 ± 0.036 | 0.974 ± 0.013 |
4.826 ± 0.029 | 5.963 ± 0.035 |
3.898 ± 0.024 | 6.586 ± 0.044 |
1.919 ± 0.018 | 6.556 ± 0.028 |
3.943 ± 0.033 | 11.003 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.798 ± 0.016 | 4.043 ± 0.03 |
4.805 ± 0.03 | 5.741 ± 0.041 |
5.507 ± 0.027 | 6.15 ± 0.025 |
5.869 ± 0.027 | 6.977 ± 0.027 |
1.42 ± 0.015 | 3.04 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
