
Schizopora paradoxa
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Hymenochaetales; Schizoporaceae; Schizopora
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
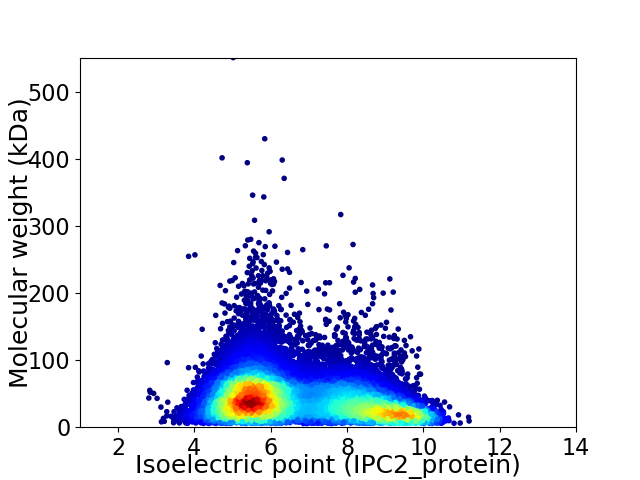
Virtual 2D-PAGE plot for 17051 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H2SJU8|A0A0H2SJU8_9AGAM Peptidase_M48 domain-containing protein OS=Schizopora paradoxa OX=27342 GN=SCHPADRAFT_937090 PE=4 SV=1
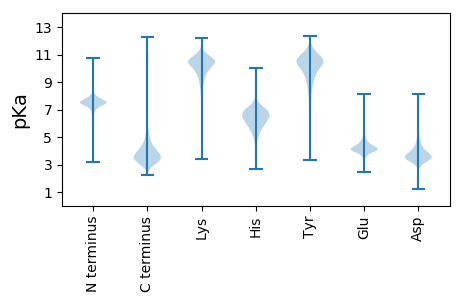
MM1 pKa = 7.03YY2 pKa = 10.56AFGGFVYY9 pKa = 10.45SLSSCPSAEE18 pKa = 3.85TMASDD23 pKa = 4.29FSTMKK28 pKa = 10.5SVGSRR33 pKa = 11.84IVVTFDD39 pKa = 2.98PCEE42 pKa = 4.57DD43 pKa = 3.91GSEE46 pKa = 3.89DD47 pKa = 3.7TYY49 pKa = 11.98DD50 pKa = 3.58NVFAAAGAAGIYY62 pKa = 8.26VIPIAWTLVDD72 pKa = 4.08SGTFTSEE79 pKa = 3.84SVPRR83 pKa = 11.84LEE85 pKa = 4.61AVTDD89 pKa = 3.93AVIKK93 pKa = 10.82NPSNVIAVAYY103 pKa = 10.08GDD105 pKa = 4.58EE106 pKa = 4.49PLLDD110 pKa = 5.2DD111 pKa = 5.48DD112 pKa = 5.94AGSASNLATYY122 pKa = 7.44VTQMKK127 pKa = 10.58SEE129 pKa = 4.07FVNAGLDD136 pKa = 3.52MPVSISEE143 pKa = 4.34MAGGWQNSANGDD155 pKa = 3.4ISSMVAAIDD164 pKa = 3.6FFMVNDD170 pKa = 3.78MPYY173 pKa = 10.02FFPDD177 pKa = 2.95ATTGGSSGSWNDD189 pKa = 3.3FVSDD193 pKa = 3.99LEE195 pKa = 4.54YY196 pKa = 10.49IASIANGKK204 pKa = 9.42HH205 pKa = 5.47IMVTQTGWPSNEE217 pKa = 4.82DD218 pKa = 3.03EE219 pKa = 4.72FAPNSNSIDD228 pKa = 3.28ASVSSEE234 pKa = 3.71AAFWQLLDD242 pKa = 3.77SHH244 pKa = 7.36CSDD247 pKa = 4.05FFKK250 pKa = 11.01PNDD253 pKa = 3.76IAWMWRR259 pKa = 11.84SWDD262 pKa = 3.53DD263 pKa = 4.98HH264 pKa = 5.12ITGWGVLDD272 pKa = 4.11SNGNPKK278 pKa = 9.09FTVQASTSCC287 pKa = 3.29
MM1 pKa = 7.03YY2 pKa = 10.56AFGGFVYY9 pKa = 10.45SLSSCPSAEE18 pKa = 3.85TMASDD23 pKa = 4.29FSTMKK28 pKa = 10.5SVGSRR33 pKa = 11.84IVVTFDD39 pKa = 2.98PCEE42 pKa = 4.57DD43 pKa = 3.91GSEE46 pKa = 3.89DD47 pKa = 3.7TYY49 pKa = 11.98DD50 pKa = 3.58NVFAAAGAAGIYY62 pKa = 8.26VIPIAWTLVDD72 pKa = 4.08SGTFTSEE79 pKa = 3.84SVPRR83 pKa = 11.84LEE85 pKa = 4.61AVTDD89 pKa = 3.93AVIKK93 pKa = 10.82NPSNVIAVAYY103 pKa = 10.08GDD105 pKa = 4.58EE106 pKa = 4.49PLLDD110 pKa = 5.2DD111 pKa = 5.48DD112 pKa = 5.94AGSASNLATYY122 pKa = 7.44VTQMKK127 pKa = 10.58SEE129 pKa = 4.07FVNAGLDD136 pKa = 3.52MPVSISEE143 pKa = 4.34MAGGWQNSANGDD155 pKa = 3.4ISSMVAAIDD164 pKa = 3.6FFMVNDD170 pKa = 3.78MPYY173 pKa = 10.02FFPDD177 pKa = 2.95ATTGGSSGSWNDD189 pKa = 3.3FVSDD193 pKa = 3.99LEE195 pKa = 4.54YY196 pKa = 10.49IASIANGKK204 pKa = 9.42HH205 pKa = 5.47IMVTQTGWPSNEE217 pKa = 4.82DD218 pKa = 3.03EE219 pKa = 4.72FAPNSNSIDD228 pKa = 3.28ASVSSEE234 pKa = 3.71AAFWQLLDD242 pKa = 3.77SHH244 pKa = 7.36CSDD247 pKa = 4.05FFKK250 pKa = 11.01PNDD253 pKa = 3.76IAWMWRR259 pKa = 11.84SWDD262 pKa = 3.53DD263 pKa = 4.98HH264 pKa = 5.12ITGWGVLDD272 pKa = 4.11SNGNPKK278 pKa = 9.09FTVQASTSCC287 pKa = 3.29
Molecular weight: 30.76 kDa
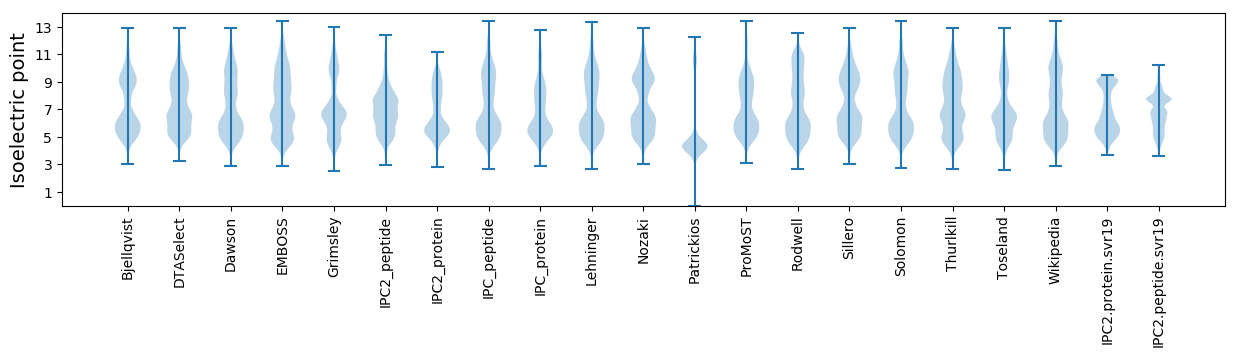
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H2RAV5|A0A0H2RAV5_9AGAM Mannose-P-dolichol utilization defect 1 protein homolog OS=Schizopora paradoxa OX=27342 GN=SCHPADRAFT_1002241 PE=3 SV=1
MM1 pKa = 7.43KK2 pKa = 10.43RR3 pKa = 11.84LLLLVVSMQVGKK15 pKa = 10.47ARR17 pKa = 11.84PSSVIRR23 pKa = 11.84RR24 pKa = 11.84PTRR27 pKa = 11.84PANLLLAFTRR37 pKa = 11.84IHH39 pKa = 6.85PPLSSRR45 pKa = 11.84RR46 pKa = 11.84EE47 pKa = 3.68RR48 pKa = 11.84ARR50 pKa = 11.84QRR52 pKa = 11.84VWRR55 pKa = 11.84MKK57 pKa = 9.91RR58 pKa = 11.84LHH60 pKa = 6.6PFGSNVRR67 pKa = 11.84PLVRR71 pKa = 11.84PLVRR75 pKa = 11.84PLVLHH80 pKa = 6.64LSPSIALLPPPSAHH94 pKa = 6.4HH95 pKa = 6.44SSPIHH100 pKa = 6.05HH101 pKa = 7.1LFITTSSRR109 pKa = 11.84ARR111 pKa = 11.84PRR113 pKa = 11.84PPKK116 pKa = 10.0ASKK119 pKa = 9.44PAPSLMPRR127 pKa = 11.84LINGSS132 pKa = 3.44
MM1 pKa = 7.43KK2 pKa = 10.43RR3 pKa = 11.84LLLLVVSMQVGKK15 pKa = 10.47ARR17 pKa = 11.84PSSVIRR23 pKa = 11.84RR24 pKa = 11.84PTRR27 pKa = 11.84PANLLLAFTRR37 pKa = 11.84IHH39 pKa = 6.85PPLSSRR45 pKa = 11.84RR46 pKa = 11.84EE47 pKa = 3.68RR48 pKa = 11.84ARR50 pKa = 11.84QRR52 pKa = 11.84VWRR55 pKa = 11.84MKK57 pKa = 9.91RR58 pKa = 11.84LHH60 pKa = 6.6PFGSNVRR67 pKa = 11.84PLVRR71 pKa = 11.84PLVRR75 pKa = 11.84PLVLHH80 pKa = 6.64LSPSIALLPPPSAHH94 pKa = 6.4HH95 pKa = 6.44SSPIHH100 pKa = 6.05HH101 pKa = 7.1LFITTSSRR109 pKa = 11.84ARR111 pKa = 11.84PRR113 pKa = 11.84PPKK116 pKa = 10.0ASKK119 pKa = 9.44PAPSLMPRR127 pKa = 11.84LINGSS132 pKa = 3.44
Molecular weight: 14.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
7036197 |
49 |
4931 |
412.7 |
45.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.975 ± 0.018 | 1.421 ± 0.008 |
5.676 ± 0.014 | 6.082 ± 0.021 |
4.121 ± 0.011 | 6.231 ± 0.015 |
2.445 ± 0.009 | 4.99 ± 0.013 |
4.746 ± 0.018 | 9.383 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.132 ± 0.007 | 3.679 ± 0.009 |
5.886 ± 0.021 | 3.501 ± 0.012 |
6.401 ± 0.017 | 9.246 ± 0.025 |
5.799 ± 0.013 | 6.341 ± 0.014 |
1.369 ± 0.007 | 2.578 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
