
Thermococcus barophilus (strain DSM 11836 / MP)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Thermococci; Thermococcales; Thermococcaceae; Thermococcus; Thermococcus barophilus
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
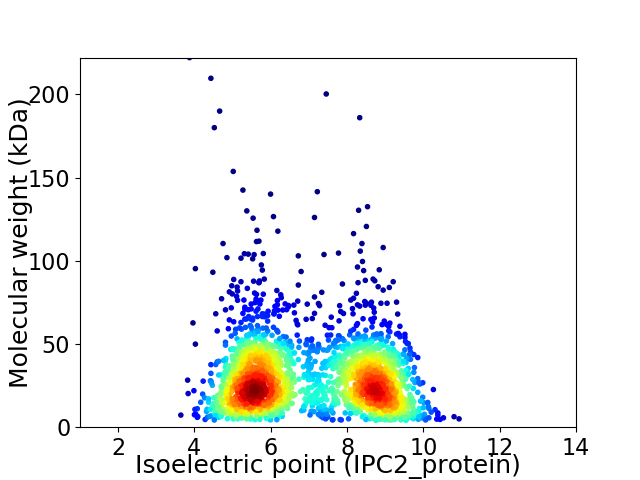
Virtual 2D-PAGE plot for 2265 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F0LJ45|F0LJ45_THEBM Threonine synthase OS=Thermococcus barophilus (strain DSM 11836 / MP) OX=391623 GN=TERMP_00414 PE=3 SV=1
MM1 pKa = 7.63KK2 pKa = 10.0VRR4 pKa = 11.84KK5 pKa = 9.01IAALAVGAAMVGATMGFASAQANLPGKK32 pKa = 10.24DD33 pKa = 3.57FFVKK37 pKa = 10.47DD38 pKa = 3.44GQPNVKK44 pKa = 9.58IVVGSQAAAMDD55 pKa = 3.88VASAADD61 pKa = 3.28IAVALGSLLYY71 pKa = 9.87TEE73 pKa = 5.42KK74 pKa = 10.36EE75 pKa = 3.84AEE77 pKa = 3.97AAGVSVVVKK86 pKa = 10.71KK87 pKa = 10.77DD88 pKa = 3.22LTPDD92 pKa = 3.1YY93 pKa = 9.82TYY95 pKa = 10.69YY96 pKa = 10.34IPVFSNYY103 pKa = 9.92YY104 pKa = 9.95EE105 pKa = 4.17DD106 pKa = 3.79TGTNPSATAWDD117 pKa = 3.8QLTDD121 pKa = 3.38NWWNGSAYY129 pKa = 10.33NGSYY133 pKa = 10.25TDD135 pKa = 3.41WTSWTPKK142 pKa = 10.24FVDD145 pKa = 3.59EE146 pKa = 4.74VEE148 pKa = 4.39NMDD151 pKa = 5.77AINGDD156 pKa = 4.04YY157 pKa = 10.36QVDD160 pKa = 3.34WDD162 pKa = 4.01FTISNIEE169 pKa = 4.14LSDD172 pKa = 3.86PEE174 pKa = 4.33QNTIVYY180 pKa = 8.87VPRR183 pKa = 11.84SADD186 pKa = 3.37LTIPAGDD193 pKa = 3.5FTVLLNYY200 pKa = 9.34TIANWTYY207 pKa = 10.9SATEE211 pKa = 4.18PDD213 pKa = 4.94SIWGTLNPSTVYY225 pKa = 11.07DD226 pKa = 4.18EE227 pKa = 5.06VHH229 pKa = 7.46DD230 pKa = 4.81DD231 pKa = 4.43DD232 pKa = 5.69NPGGYY237 pKa = 7.79TFSGYY242 pKa = 10.14IYY244 pKa = 10.06DD245 pKa = 4.04GVGAGDD251 pKa = 3.82TFTVLGNTYY260 pKa = 10.58YY261 pKa = 10.33ILDD264 pKa = 3.81VLADD268 pKa = 4.76GIKK271 pKa = 10.1YY272 pKa = 10.14GHH274 pKa = 7.05DD275 pKa = 3.21HH276 pKa = 5.57GQVWFHH282 pKa = 6.83VGDD285 pKa = 3.6VKK287 pKa = 11.17EE288 pKa = 3.93FDD290 pKa = 3.67GYY292 pKa = 10.7KK293 pKa = 10.0IKK295 pKa = 10.88AVDD298 pKa = 3.47ISVSPSNKK306 pKa = 9.8ALFEE310 pKa = 3.76ITAPDD315 pKa = 3.69GRR317 pKa = 11.84SDD319 pKa = 4.05LVIISTDD326 pKa = 3.26EE327 pKa = 4.21GDD329 pKa = 3.33VDD331 pKa = 4.87ISTKK335 pKa = 9.65SDD337 pKa = 2.83KK338 pKa = 10.74FNPGEE343 pKa = 4.21VVLKK347 pKa = 10.9LDD349 pKa = 3.76DD350 pKa = 3.92TFVGIDD356 pKa = 3.62GNLIAQLEE364 pKa = 4.37VRR366 pKa = 11.84TNVVEE371 pKa = 3.99VHH373 pKa = 6.15TGDD376 pKa = 3.73EE377 pKa = 4.49LVSGWVVNFTIEE389 pKa = 4.03NNKK392 pKa = 9.3VKK394 pKa = 10.2WLTLTNANDD403 pKa = 3.96LSGSTLDD410 pKa = 3.71ILGKK414 pKa = 10.01YY415 pKa = 8.66KK416 pKa = 9.95MYY418 pKa = 11.15YY419 pKa = 9.96EE420 pKa = 4.4VDD422 pKa = 3.21SHH424 pKa = 6.14TLEE427 pKa = 4.57TDD429 pKa = 2.96DD430 pKa = 3.56TTYY433 pKa = 11.16YY434 pKa = 10.24AAKK437 pKa = 9.97AYY439 pKa = 9.66IVVKK443 pKa = 9.69PSEE446 pKa = 4.62PIIQTEE452 pKa = 3.99EE453 pKa = 4.13LKK455 pKa = 11.37VGDD458 pKa = 4.32EE459 pKa = 4.26VSDD462 pKa = 3.72TGWVVDD468 pKa = 4.06QIKK471 pKa = 10.54GGTYY475 pKa = 8.91TEE477 pKa = 4.22VTVMHH482 pKa = 6.26PTEE485 pKa = 5.31PITYY489 pKa = 9.33LDD491 pKa = 3.81TEE493 pKa = 4.56IDD495 pKa = 3.75PEE497 pKa = 4.99NIDD500 pKa = 3.59SNLILVGGPVANAVTKK516 pKa = 10.22YY517 pKa = 10.65LVDD520 pKa = 3.41NGYY523 pKa = 8.26STVDD527 pKa = 3.22WYY529 pKa = 11.79NSAGDD534 pKa = 3.36IEE536 pKa = 4.85YY537 pKa = 10.29IEE539 pKa = 5.35DD540 pKa = 3.69YY541 pKa = 11.02NGFGILIVAGKK552 pKa = 10.0DD553 pKa = 3.02RR554 pKa = 11.84YY555 pKa = 8.98ATRR558 pKa = 11.84EE559 pKa = 3.69AAKK562 pKa = 10.6QLMEE566 pKa = 4.0YY567 pKa = 10.18LANLSS572 pKa = 3.58
MM1 pKa = 7.63KK2 pKa = 10.0VRR4 pKa = 11.84KK5 pKa = 9.01IAALAVGAAMVGATMGFASAQANLPGKK32 pKa = 10.24DD33 pKa = 3.57FFVKK37 pKa = 10.47DD38 pKa = 3.44GQPNVKK44 pKa = 9.58IVVGSQAAAMDD55 pKa = 3.88VASAADD61 pKa = 3.28IAVALGSLLYY71 pKa = 9.87TEE73 pKa = 5.42KK74 pKa = 10.36EE75 pKa = 3.84AEE77 pKa = 3.97AAGVSVVVKK86 pKa = 10.71KK87 pKa = 10.77DD88 pKa = 3.22LTPDD92 pKa = 3.1YY93 pKa = 9.82TYY95 pKa = 10.69YY96 pKa = 10.34IPVFSNYY103 pKa = 9.92YY104 pKa = 9.95EE105 pKa = 4.17DD106 pKa = 3.79TGTNPSATAWDD117 pKa = 3.8QLTDD121 pKa = 3.38NWWNGSAYY129 pKa = 10.33NGSYY133 pKa = 10.25TDD135 pKa = 3.41WTSWTPKK142 pKa = 10.24FVDD145 pKa = 3.59EE146 pKa = 4.74VEE148 pKa = 4.39NMDD151 pKa = 5.77AINGDD156 pKa = 4.04YY157 pKa = 10.36QVDD160 pKa = 3.34WDD162 pKa = 4.01FTISNIEE169 pKa = 4.14LSDD172 pKa = 3.86PEE174 pKa = 4.33QNTIVYY180 pKa = 8.87VPRR183 pKa = 11.84SADD186 pKa = 3.37LTIPAGDD193 pKa = 3.5FTVLLNYY200 pKa = 9.34TIANWTYY207 pKa = 10.9SATEE211 pKa = 4.18PDD213 pKa = 4.94SIWGTLNPSTVYY225 pKa = 11.07DD226 pKa = 4.18EE227 pKa = 5.06VHH229 pKa = 7.46DD230 pKa = 4.81DD231 pKa = 4.43DD232 pKa = 5.69NPGGYY237 pKa = 7.79TFSGYY242 pKa = 10.14IYY244 pKa = 10.06DD245 pKa = 4.04GVGAGDD251 pKa = 3.82TFTVLGNTYY260 pKa = 10.58YY261 pKa = 10.33ILDD264 pKa = 3.81VLADD268 pKa = 4.76GIKK271 pKa = 10.1YY272 pKa = 10.14GHH274 pKa = 7.05DD275 pKa = 3.21HH276 pKa = 5.57GQVWFHH282 pKa = 6.83VGDD285 pKa = 3.6VKK287 pKa = 11.17EE288 pKa = 3.93FDD290 pKa = 3.67GYY292 pKa = 10.7KK293 pKa = 10.0IKK295 pKa = 10.88AVDD298 pKa = 3.47ISVSPSNKK306 pKa = 9.8ALFEE310 pKa = 3.76ITAPDD315 pKa = 3.69GRR317 pKa = 11.84SDD319 pKa = 4.05LVIISTDD326 pKa = 3.26EE327 pKa = 4.21GDD329 pKa = 3.33VDD331 pKa = 4.87ISTKK335 pKa = 9.65SDD337 pKa = 2.83KK338 pKa = 10.74FNPGEE343 pKa = 4.21VVLKK347 pKa = 10.9LDD349 pKa = 3.76DD350 pKa = 3.92TFVGIDD356 pKa = 3.62GNLIAQLEE364 pKa = 4.37VRR366 pKa = 11.84TNVVEE371 pKa = 3.99VHH373 pKa = 6.15TGDD376 pKa = 3.73EE377 pKa = 4.49LVSGWVVNFTIEE389 pKa = 4.03NNKK392 pKa = 9.3VKK394 pKa = 10.2WLTLTNANDD403 pKa = 3.96LSGSTLDD410 pKa = 3.71ILGKK414 pKa = 10.01YY415 pKa = 8.66KK416 pKa = 9.95MYY418 pKa = 11.15YY419 pKa = 9.96EE420 pKa = 4.4VDD422 pKa = 3.21SHH424 pKa = 6.14TLEE427 pKa = 4.57TDD429 pKa = 2.96DD430 pKa = 3.56TTYY433 pKa = 11.16YY434 pKa = 10.24AAKK437 pKa = 9.97AYY439 pKa = 9.66IVVKK443 pKa = 9.69PSEE446 pKa = 4.62PIIQTEE452 pKa = 3.99EE453 pKa = 4.13LKK455 pKa = 11.37VGDD458 pKa = 4.32EE459 pKa = 4.26VSDD462 pKa = 3.72TGWVVDD468 pKa = 4.06QIKK471 pKa = 10.54GGTYY475 pKa = 8.91TEE477 pKa = 4.22VTVMHH482 pKa = 6.26PTEE485 pKa = 5.31PITYY489 pKa = 9.33LDD491 pKa = 3.81TEE493 pKa = 4.56IDD495 pKa = 3.75PEE497 pKa = 4.99NIDD500 pKa = 3.59SNLILVGGPVANAVTKK516 pKa = 10.22YY517 pKa = 10.65LVDD520 pKa = 3.41NGYY523 pKa = 8.26STVDD527 pKa = 3.22WYY529 pKa = 11.79NSAGDD534 pKa = 3.36IEE536 pKa = 4.85YY537 pKa = 10.29IEE539 pKa = 5.35DD540 pKa = 3.69YY541 pKa = 11.02NGFGILIVAGKK552 pKa = 10.0DD553 pKa = 3.02RR554 pKa = 11.84YY555 pKa = 8.98ATRR558 pKa = 11.84EE559 pKa = 3.69AAKK562 pKa = 10.6QLMEE566 pKa = 4.0YY567 pKa = 10.18LANLSS572 pKa = 3.58
Molecular weight: 62.65 kDa
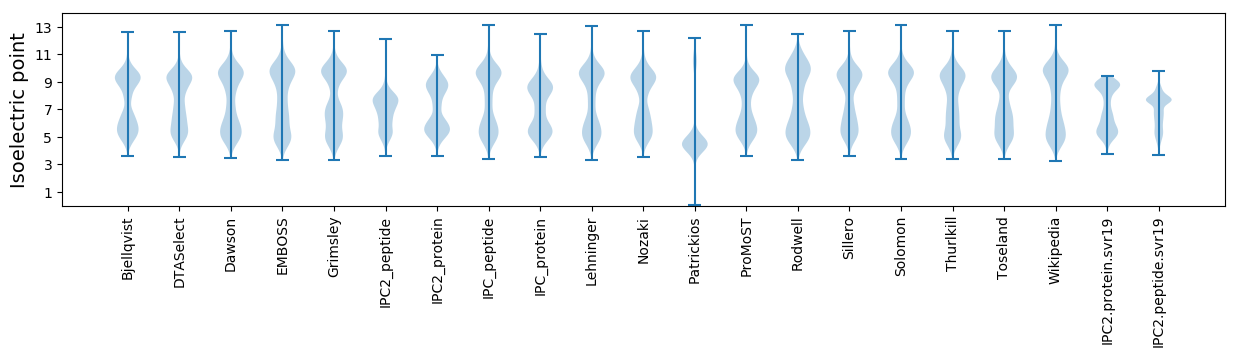
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F0LJS1|F0LJS1_THEBM Peptidase_M28 domain-containing protein OS=Thermococcus barophilus (strain DSM 11836 / MP) OX=391623 GN=TERMP_01738 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.23RR3 pKa = 11.84RR4 pKa = 11.84PRR6 pKa = 11.84KK7 pKa = 7.59WKK9 pKa = 10.02KK10 pKa = 8.58KK11 pKa = 10.27GRR13 pKa = 11.84MRR15 pKa = 11.84WKK17 pKa = 9.51WIKK20 pKa = 10.46KK21 pKa = 8.96RR22 pKa = 11.84IRR24 pKa = 11.84RR25 pKa = 11.84LKK27 pKa = 9.33RR28 pKa = 11.84QRR30 pKa = 11.84KK31 pKa = 8.4KK32 pKa = 10.86EE33 pKa = 3.77RR34 pKa = 11.84GLII37 pKa = 3.84
MM1 pKa = 7.48KK2 pKa = 10.23RR3 pKa = 11.84RR4 pKa = 11.84PRR6 pKa = 11.84KK7 pKa = 7.59WKK9 pKa = 10.02KK10 pKa = 8.58KK11 pKa = 10.27GRR13 pKa = 11.84MRR15 pKa = 11.84WKK17 pKa = 9.51WIKK20 pKa = 10.46KK21 pKa = 8.96RR22 pKa = 11.84IRR24 pKa = 11.84RR25 pKa = 11.84LKK27 pKa = 9.33RR28 pKa = 11.84QRR30 pKa = 11.84KK31 pKa = 8.4KK32 pKa = 10.86EE33 pKa = 3.77RR34 pKa = 11.84GLII37 pKa = 3.84
Molecular weight: 5.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
631540 |
37 |
2030 |
278.8 |
31.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.876 ± 0.051 | 0.585 ± 0.018 |
4.523 ± 0.035 | 8.691 ± 0.075 |
4.617 ± 0.042 | 6.967 ± 0.048 |
1.613 ± 0.02 | 8.589 ± 0.045 |
8.368 ± 0.072 | 10.301 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.344 ± 0.025 | 3.512 ± 0.043 |
4.064 ± 0.035 | 1.969 ± 0.027 |
5.049 ± 0.044 | 4.823 ± 0.046 |
4.356 ± 0.046 | 7.497 ± 0.044 |
1.187 ± 0.019 | 4.069 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
