
Sea otter polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0S3KGJ2|A0A0S3KGJ2_9POLY Capsid protein VP1 OS=Sea otter polyomavirus 1 OX=1552409 PE=3 SV=1
MM1 pKa = 7.57DD2 pKa = 3.91TVLSRR7 pKa = 11.84EE8 pKa = 4.3DD9 pKa = 3.25ARR11 pKa = 11.84EE12 pKa = 3.79LMGLLRR18 pKa = 11.84LDD20 pKa = 3.24MAVYY24 pKa = 10.77GNLPLMRR31 pKa = 11.84KK32 pKa = 9.6AYY34 pKa = 10.35LKK36 pKa = 10.23RR37 pKa = 11.84CKK39 pKa = 9.66EE40 pKa = 3.83LHH42 pKa = 6.38PDD44 pKa = 3.2KK45 pKa = 11.49GGDD48 pKa = 3.17EE49 pKa = 4.3TMMKK53 pKa = 10.2KK54 pKa = 10.58LNEE57 pKa = 4.17LYY59 pKa = 10.96KK60 pKa = 11.07LMEE63 pKa = 4.78DD64 pKa = 4.42NIRR67 pKa = 11.84EE68 pKa = 3.99VHH70 pKa = 6.5SNVEE74 pKa = 3.76AAGWKK79 pKa = 9.99AEE81 pKa = 3.94EE82 pKa = 4.38VPTYY86 pKa = 9.61GTTEE90 pKa = 3.86WEE92 pKa = 4.4HH93 pKa = 4.47WWEE96 pKa = 3.99AFNANWNDD104 pKa = 3.33LRR106 pKa = 11.84CDD108 pKa = 3.58EE109 pKa = 4.46EE110 pKa = 5.81LEE112 pKa = 4.69EE113 pKa = 4.8EE114 pKa = 4.84DD115 pKa = 5.88LDD117 pKa = 4.98DD118 pKa = 4.68SQHH121 pKa = 5.37STPPKK126 pKa = 8.79KK127 pKa = 9.9KK128 pKa = 10.08RR129 pKa = 11.84RR130 pKa = 11.84GDD132 pKa = 3.49EE133 pKa = 3.96PKK135 pKa = 10.66DD136 pKa = 3.66FPEE139 pKa = 4.27SLRR142 pKa = 11.84TMLSQAIFSNRR153 pKa = 11.84TLSCFAIYY161 pKa = 7.53TTSDD165 pKa = 2.95KK166 pKa = 11.34ARR168 pKa = 11.84LLFKK172 pKa = 10.96KK173 pKa = 10.73LMDD176 pKa = 4.02KK177 pKa = 11.06YY178 pKa = 10.36SANFISLHH186 pKa = 5.37SCGIHH191 pKa = 6.97SILFLLTPQKK201 pKa = 10.8HH202 pKa = 4.9RR203 pKa = 11.84VSAINNFCLKK213 pKa = 10.2FCTISFLICKK223 pKa = 9.0GVNKK227 pKa = 9.74EE228 pKa = 3.81YY229 pKa = 10.87CLYY232 pKa = 10.72KK233 pKa = 10.61ALCTDD238 pKa = 4.1PFKK241 pKa = 11.12LLQEE245 pKa = 4.31SVEE248 pKa = 4.44GGLSEE253 pKa = 4.42NYY255 pKa = 9.87FMPEE259 pKa = 3.32EE260 pKa = 4.38GDD262 pKa = 3.65EE263 pKa = 4.28VKK265 pKa = 10.54QVSWKK270 pKa = 9.99MITEE274 pKa = 4.03YY275 pKa = 10.8AVEE278 pKa = 4.17IDD280 pKa = 5.24CDD282 pKa = 3.94DD283 pKa = 4.92LYY285 pKa = 11.99LLMGLYY291 pKa = 10.33LEE293 pKa = 5.04YY294 pKa = 9.9ATEE297 pKa = 4.01PDD299 pKa = 3.31YY300 pKa = 11.76CKK302 pKa = 10.18ICSSKK307 pKa = 10.62SIPVHH312 pKa = 5.02YY313 pKa = 9.87RR314 pKa = 11.84NHH316 pKa = 6.39GKK318 pKa = 8.39HH319 pKa = 6.35HH320 pKa = 6.36GNALLFLEE328 pKa = 4.99SKK330 pKa = 9.11NQKK333 pKa = 9.97AICQQAVDD341 pKa = 3.66TVIAKK346 pKa = 10.02KK347 pKa = 10.28RR348 pKa = 11.84VEE350 pKa = 4.11TKK352 pKa = 9.65YY353 pKa = 10.85LSRR356 pKa = 11.84ADD358 pKa = 3.55MLTKK362 pKa = 10.35RR363 pKa = 11.84FHH365 pKa = 6.89KK366 pKa = 10.3LLDD369 pKa = 3.34MMEE372 pKa = 4.63DD373 pKa = 3.3MFGASGSADD382 pKa = 3.41LKK384 pKa = 11.33LYY386 pKa = 9.83MCGCAWLEE394 pKa = 3.79CLQYY398 pKa = 11.17CMEE401 pKa = 4.22SVVYY405 pKa = 10.04EE406 pKa = 4.55FLRR409 pKa = 11.84TMVTNIPKK417 pKa = 9.67KK418 pKa = 10.39RR419 pKa = 11.84YY420 pKa = 8.08WLFKK424 pKa = 10.95GPIDD428 pKa = 3.83TGKK431 pKa = 7.19TTLAAGLLNLCGGRR445 pKa = 11.84ALNINLPQDD454 pKa = 3.49RR455 pKa = 11.84LPFEE459 pKa = 4.86LGVAIDD465 pKa = 3.29QFMVVFEE472 pKa = 4.44DD473 pKa = 3.83VKK475 pKa = 11.04GHH477 pKa = 6.51GGRR480 pKa = 11.84DD481 pKa = 3.59LPPGQGVHH489 pKa = 6.25NLDD492 pKa = 3.49NLRR495 pKa = 11.84DD496 pKa = 3.88YY497 pKa = 11.6LDD499 pKa = 4.13GSVKK503 pKa = 10.37VNLEE507 pKa = 3.85KK508 pKa = 10.85KK509 pKa = 9.59HH510 pKa = 5.0VNKK513 pKa = 9.3RR514 pKa = 11.84TQIFPPGLVTMNEE527 pKa = 3.5YY528 pKa = 10.7DD529 pKa = 3.42IPRR532 pKa = 11.84TLLARR537 pKa = 11.84FARR540 pKa = 11.84IVEE543 pKa = 4.37FRR545 pKa = 11.84PKK547 pKa = 9.9PHH549 pKa = 7.36LKK551 pKa = 10.19RR552 pKa = 11.84SLNNTPEE559 pKa = 4.33LLEE562 pKa = 3.85QRR564 pKa = 11.84IVQSGVTLLLLLIWHH579 pKa = 6.65RR580 pKa = 11.84PVYY583 pKa = 10.83DD584 pKa = 3.69FDD586 pKa = 4.85EE587 pKa = 5.68DD588 pKa = 3.22IQARR592 pKa = 11.84VVEE595 pKa = 4.53WKK597 pKa = 10.15EE598 pKa = 3.82RR599 pKa = 11.84LDD601 pKa = 4.13SEE603 pKa = 5.15IPCTMYY609 pKa = 11.07SRR611 pKa = 11.84MKK613 pKa = 10.59RR614 pKa = 11.84NVACGVHH621 pKa = 6.06ILSDD625 pKa = 3.46TADD628 pKa = 3.38EE629 pKa = 4.62GRR631 pKa = 11.84AFRR634 pKa = 11.84PQPPPPPQPADD645 pKa = 3.63DD646 pKa = 4.55SEE648 pKa = 6.05ADD650 pKa = 3.42SGMDD654 pKa = 3.33SARR657 pKa = 11.84SQSSSEE663 pKa = 4.09DD664 pKa = 3.5TDD666 pKa = 4.97DD667 pKa = 4.3GFPPVPPPSPVPFF680 pKa = 4.65
MM1 pKa = 7.57DD2 pKa = 3.91TVLSRR7 pKa = 11.84EE8 pKa = 4.3DD9 pKa = 3.25ARR11 pKa = 11.84EE12 pKa = 3.79LMGLLRR18 pKa = 11.84LDD20 pKa = 3.24MAVYY24 pKa = 10.77GNLPLMRR31 pKa = 11.84KK32 pKa = 9.6AYY34 pKa = 10.35LKK36 pKa = 10.23RR37 pKa = 11.84CKK39 pKa = 9.66EE40 pKa = 3.83LHH42 pKa = 6.38PDD44 pKa = 3.2KK45 pKa = 11.49GGDD48 pKa = 3.17EE49 pKa = 4.3TMMKK53 pKa = 10.2KK54 pKa = 10.58LNEE57 pKa = 4.17LYY59 pKa = 10.96KK60 pKa = 11.07LMEE63 pKa = 4.78DD64 pKa = 4.42NIRR67 pKa = 11.84EE68 pKa = 3.99VHH70 pKa = 6.5SNVEE74 pKa = 3.76AAGWKK79 pKa = 9.99AEE81 pKa = 3.94EE82 pKa = 4.38VPTYY86 pKa = 9.61GTTEE90 pKa = 3.86WEE92 pKa = 4.4HH93 pKa = 4.47WWEE96 pKa = 3.99AFNANWNDD104 pKa = 3.33LRR106 pKa = 11.84CDD108 pKa = 3.58EE109 pKa = 4.46EE110 pKa = 5.81LEE112 pKa = 4.69EE113 pKa = 4.8EE114 pKa = 4.84DD115 pKa = 5.88LDD117 pKa = 4.98DD118 pKa = 4.68SQHH121 pKa = 5.37STPPKK126 pKa = 8.79KK127 pKa = 9.9KK128 pKa = 10.08RR129 pKa = 11.84RR130 pKa = 11.84GDD132 pKa = 3.49EE133 pKa = 3.96PKK135 pKa = 10.66DD136 pKa = 3.66FPEE139 pKa = 4.27SLRR142 pKa = 11.84TMLSQAIFSNRR153 pKa = 11.84TLSCFAIYY161 pKa = 7.53TTSDD165 pKa = 2.95KK166 pKa = 11.34ARR168 pKa = 11.84LLFKK172 pKa = 10.96KK173 pKa = 10.73LMDD176 pKa = 4.02KK177 pKa = 11.06YY178 pKa = 10.36SANFISLHH186 pKa = 5.37SCGIHH191 pKa = 6.97SILFLLTPQKK201 pKa = 10.8HH202 pKa = 4.9RR203 pKa = 11.84VSAINNFCLKK213 pKa = 10.2FCTISFLICKK223 pKa = 9.0GVNKK227 pKa = 9.74EE228 pKa = 3.81YY229 pKa = 10.87CLYY232 pKa = 10.72KK233 pKa = 10.61ALCTDD238 pKa = 4.1PFKK241 pKa = 11.12LLQEE245 pKa = 4.31SVEE248 pKa = 4.44GGLSEE253 pKa = 4.42NYY255 pKa = 9.87FMPEE259 pKa = 3.32EE260 pKa = 4.38GDD262 pKa = 3.65EE263 pKa = 4.28VKK265 pKa = 10.54QVSWKK270 pKa = 9.99MITEE274 pKa = 4.03YY275 pKa = 10.8AVEE278 pKa = 4.17IDD280 pKa = 5.24CDD282 pKa = 3.94DD283 pKa = 4.92LYY285 pKa = 11.99LLMGLYY291 pKa = 10.33LEE293 pKa = 5.04YY294 pKa = 9.9ATEE297 pKa = 4.01PDD299 pKa = 3.31YY300 pKa = 11.76CKK302 pKa = 10.18ICSSKK307 pKa = 10.62SIPVHH312 pKa = 5.02YY313 pKa = 9.87RR314 pKa = 11.84NHH316 pKa = 6.39GKK318 pKa = 8.39HH319 pKa = 6.35HH320 pKa = 6.36GNALLFLEE328 pKa = 4.99SKK330 pKa = 9.11NQKK333 pKa = 9.97AICQQAVDD341 pKa = 3.66TVIAKK346 pKa = 10.02KK347 pKa = 10.28RR348 pKa = 11.84VEE350 pKa = 4.11TKK352 pKa = 9.65YY353 pKa = 10.85LSRR356 pKa = 11.84ADD358 pKa = 3.55MLTKK362 pKa = 10.35RR363 pKa = 11.84FHH365 pKa = 6.89KK366 pKa = 10.3LLDD369 pKa = 3.34MMEE372 pKa = 4.63DD373 pKa = 3.3MFGASGSADD382 pKa = 3.41LKK384 pKa = 11.33LYY386 pKa = 9.83MCGCAWLEE394 pKa = 3.79CLQYY398 pKa = 11.17CMEE401 pKa = 4.22SVVYY405 pKa = 10.04EE406 pKa = 4.55FLRR409 pKa = 11.84TMVTNIPKK417 pKa = 9.67KK418 pKa = 10.39RR419 pKa = 11.84YY420 pKa = 8.08WLFKK424 pKa = 10.95GPIDD428 pKa = 3.83TGKK431 pKa = 7.19TTLAAGLLNLCGGRR445 pKa = 11.84ALNINLPQDD454 pKa = 3.49RR455 pKa = 11.84LPFEE459 pKa = 4.86LGVAIDD465 pKa = 3.29QFMVVFEE472 pKa = 4.44DD473 pKa = 3.83VKK475 pKa = 11.04GHH477 pKa = 6.51GGRR480 pKa = 11.84DD481 pKa = 3.59LPPGQGVHH489 pKa = 6.25NLDD492 pKa = 3.49NLRR495 pKa = 11.84DD496 pKa = 3.88YY497 pKa = 11.6LDD499 pKa = 4.13GSVKK503 pKa = 10.37VNLEE507 pKa = 3.85KK508 pKa = 10.85KK509 pKa = 9.59HH510 pKa = 5.0VNKK513 pKa = 9.3RR514 pKa = 11.84TQIFPPGLVTMNEE527 pKa = 3.5YY528 pKa = 10.7DD529 pKa = 3.42IPRR532 pKa = 11.84TLLARR537 pKa = 11.84FARR540 pKa = 11.84IVEE543 pKa = 4.37FRR545 pKa = 11.84PKK547 pKa = 9.9PHH549 pKa = 7.36LKK551 pKa = 10.19RR552 pKa = 11.84SLNNTPEE559 pKa = 4.33LLEE562 pKa = 3.85QRR564 pKa = 11.84IVQSGVTLLLLLIWHH579 pKa = 6.65RR580 pKa = 11.84PVYY583 pKa = 10.83DD584 pKa = 3.69FDD586 pKa = 4.85EE587 pKa = 5.68DD588 pKa = 3.22IQARR592 pKa = 11.84VVEE595 pKa = 4.53WKK597 pKa = 10.15EE598 pKa = 3.82RR599 pKa = 11.84LDD601 pKa = 4.13SEE603 pKa = 5.15IPCTMYY609 pKa = 11.07SRR611 pKa = 11.84MKK613 pKa = 10.59RR614 pKa = 11.84NVACGVHH621 pKa = 6.06ILSDD625 pKa = 3.46TADD628 pKa = 3.38EE629 pKa = 4.62GRR631 pKa = 11.84AFRR634 pKa = 11.84PQPPPPPQPADD645 pKa = 3.63DD646 pKa = 4.55SEE648 pKa = 6.05ADD650 pKa = 3.42SGMDD654 pKa = 3.33SARR657 pKa = 11.84SQSSSEE663 pKa = 4.09DD664 pKa = 3.5TDD666 pKa = 4.97DD667 pKa = 4.3GFPPVPPPSPVPFF680 pKa = 4.65
Molecular weight: 77.9 kDa
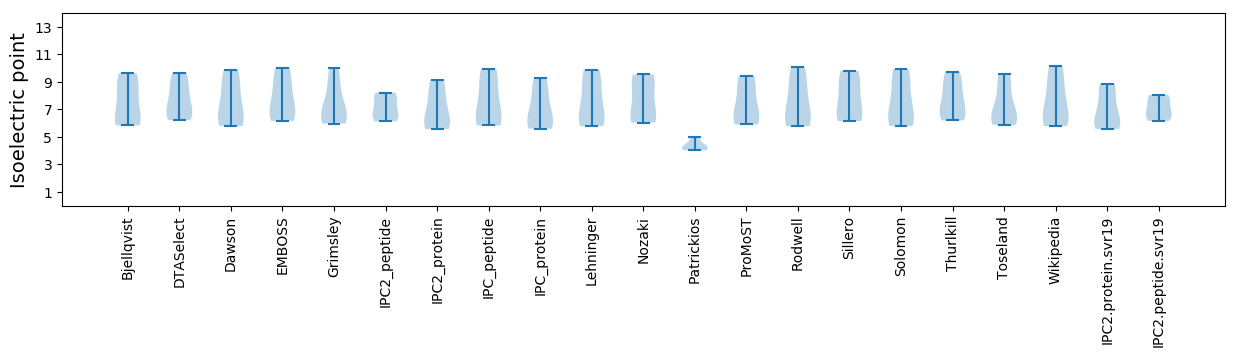
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A097A5I9|A0A097A5I9_9POLY Large T antigen OS=Sea otter polyomavirus 1 OX=1552409 PE=4 SV=1
MM1 pKa = 7.69ALQVWYY7 pKa = 9.71PDD9 pKa = 3.57FQEE12 pKa = 4.03MLFPGVNSLVRR23 pKa = 11.84ALHH26 pKa = 5.96YY27 pKa = 10.16IDD29 pKa = 3.85PRR31 pKa = 11.84EE32 pKa = 4.07WAASLYY38 pKa = 10.7AQLRR42 pKa = 11.84DD43 pKa = 3.55LFGYY47 pKa = 10.3GPARR51 pKa = 11.84LGYY54 pKa = 8.33STTEE58 pKa = 3.7VGQATGISFRR68 pKa = 11.84DD69 pKa = 3.48SVARR73 pKa = 11.84LLEE76 pKa = 4.19EE77 pKa = 4.14VTWAVSSAPLRR88 pKa = 11.84TYY90 pKa = 10.9QFLADD95 pKa = 3.95YY96 pKa = 9.94YY97 pKa = 11.24SSLSHH102 pKa = 6.45IRR104 pKa = 11.84PSMVRR109 pKa = 11.84QIAQSHH115 pKa = 6.3GPLPSKK121 pKa = 10.73FIDD124 pKa = 3.94YY125 pKa = 10.8KK126 pKa = 11.39NLDD129 pKa = 3.67NGPFSGEE136 pKa = 4.28LVDD139 pKa = 5.18KK140 pKa = 10.93APPPGGANQRR150 pKa = 11.84VTPDD154 pKa = 2.78WLLPLILDD162 pKa = 4.76LYY164 pKa = 11.14GPLNPSWKK172 pKa = 10.11EE173 pKa = 3.86EE174 pKa = 4.09EE175 pKa = 4.87DD176 pKa = 4.49DD177 pKa = 4.58GPQKK181 pKa = 10.63KK182 pKa = 8.96IQRR185 pKa = 11.84TNTRR189 pKa = 11.84AKK191 pKa = 10.61APNKK195 pKa = 9.29RR196 pKa = 11.84RR197 pKa = 11.84NRR199 pKa = 11.84GSGGQKK205 pKa = 9.46RR206 pKa = 11.84ARR208 pKa = 11.84KK209 pKa = 9.41HH210 pKa = 5.19SRR212 pKa = 11.84NN213 pKa = 3.5
MM1 pKa = 7.69ALQVWYY7 pKa = 9.71PDD9 pKa = 3.57FQEE12 pKa = 4.03MLFPGVNSLVRR23 pKa = 11.84ALHH26 pKa = 5.96YY27 pKa = 10.16IDD29 pKa = 3.85PRR31 pKa = 11.84EE32 pKa = 4.07WAASLYY38 pKa = 10.7AQLRR42 pKa = 11.84DD43 pKa = 3.55LFGYY47 pKa = 10.3GPARR51 pKa = 11.84LGYY54 pKa = 8.33STTEE58 pKa = 3.7VGQATGISFRR68 pKa = 11.84DD69 pKa = 3.48SVARR73 pKa = 11.84LLEE76 pKa = 4.19EE77 pKa = 4.14VTWAVSSAPLRR88 pKa = 11.84TYY90 pKa = 10.9QFLADD95 pKa = 3.95YY96 pKa = 9.94YY97 pKa = 11.24SSLSHH102 pKa = 6.45IRR104 pKa = 11.84PSMVRR109 pKa = 11.84QIAQSHH115 pKa = 6.3GPLPSKK121 pKa = 10.73FIDD124 pKa = 3.94YY125 pKa = 10.8KK126 pKa = 11.39NLDD129 pKa = 3.67NGPFSGEE136 pKa = 4.28LVDD139 pKa = 5.18KK140 pKa = 10.93APPPGGANQRR150 pKa = 11.84VTPDD154 pKa = 2.78WLLPLILDD162 pKa = 4.76LYY164 pKa = 11.14GPLNPSWKK172 pKa = 10.11EE173 pKa = 3.86EE174 pKa = 4.09EE175 pKa = 4.87DD176 pKa = 4.49DD177 pKa = 4.58GPQKK181 pKa = 10.63KK182 pKa = 8.96IQRR185 pKa = 11.84TNTRR189 pKa = 11.84AKK191 pKa = 10.61APNKK195 pKa = 9.29RR196 pKa = 11.84RR197 pKa = 11.84NRR199 pKa = 11.84GSGGQKK205 pKa = 9.46RR206 pKa = 11.84ARR208 pKa = 11.84KK209 pKa = 9.41HH210 pKa = 5.19SRR212 pKa = 11.84NN213 pKa = 3.5
Molecular weight: 24.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1823 |
69 |
680 |
303.8 |
34.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.308 ± 1.362 | 2.139 ± 0.742 |
5.979 ± 0.543 | 6.363 ± 0.564 |
3.511 ± 0.235 | 7.076 ± 0.966 |
2.194 ± 0.222 | 4.059 ± 0.332 |
5.979 ± 0.795 | 10.697 ± 0.568 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.852 ± 0.581 | 4.114 ± 0.197 |
6.418 ± 0.701 | 3.62 ± 0.778 |
6.089 ± 0.436 | 6.473 ± 0.614 |
4.882 ± 0.367 | 5.595 ± 0.412 |
1.81 ± 0.308 | 3.84 ± 0.225 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
