
Drosophila immigrans Nora virus
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
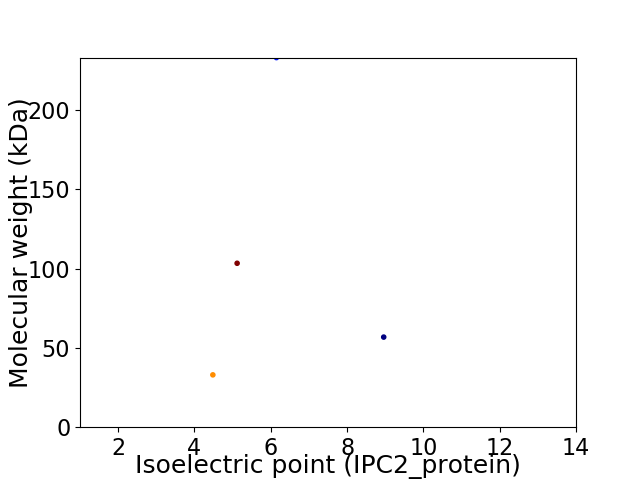
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075E230|A0A075E230_9VIRU VP1 OS=Drosophila immigrans Nora virus OX=1500866 GN=VP1 PE=4 SV=1
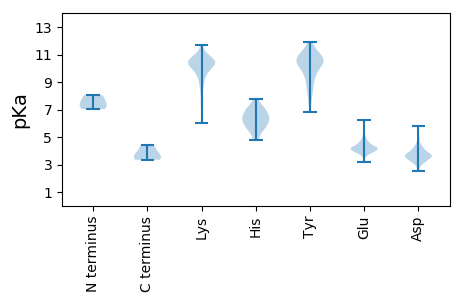
MM1 pKa = 7.65ALKK4 pKa = 10.46QEE6 pKa = 4.1IFEE9 pKa = 4.34QNSTLFEE16 pKa = 4.1VLDD19 pKa = 4.11EE20 pKa = 4.47NEE22 pKa = 3.8VMEE25 pKa = 4.51IKK27 pKa = 10.3EE28 pKa = 4.12IQSSVTAVQTQVDD41 pKa = 4.02QQKK44 pKa = 10.33LQLDD48 pKa = 4.17GLARR52 pKa = 11.84RR53 pKa = 11.84VDD55 pKa = 3.92LNQARR60 pKa = 11.84NEE62 pKa = 4.02EE63 pKa = 4.13QFVNFNTMLIATNTEE78 pKa = 3.38VDD80 pKa = 3.5KK81 pKa = 11.63LKK83 pKa = 8.78TTTTTLSVSVNNLEE97 pKa = 4.06RR98 pKa = 11.84SVGEE102 pKa = 4.62LEE104 pKa = 4.22TTTQSNFDD112 pKa = 3.8TIFRR116 pKa = 11.84RR117 pKa = 11.84LAITEE122 pKa = 4.19EE123 pKa = 4.19TSSQNKK129 pKa = 9.92SEE131 pKa = 4.09IVTILDD137 pKa = 4.13KK138 pKa = 10.85IDD140 pKa = 4.31DD141 pKa = 4.41LEE143 pKa = 5.5HH144 pKa = 7.26SVEE147 pKa = 4.21SANDD151 pKa = 3.63EE152 pKa = 4.16IAEE155 pKa = 4.22LTDD158 pKa = 3.84SVEE161 pKa = 4.47DD162 pKa = 3.61NQTAITTLSGRR173 pKa = 11.84VTTLADD179 pKa = 3.11RR180 pKa = 11.84VTKK183 pKa = 10.78LEE185 pKa = 4.7NIVQSISPYY194 pKa = 8.79IQRR197 pKa = 11.84QTFYY201 pKa = 10.87RR202 pKa = 11.84IVYY205 pKa = 8.38YY206 pKa = 9.05KK207 pKa = 10.69TNGSMRR213 pKa = 11.84NVGFWANGYY222 pKa = 8.78VRR224 pKa = 11.84YY225 pKa = 10.65SMSFQVGTSATANFFTNLTITSVISSYY252 pKa = 11.14DD253 pKa = 3.35YY254 pKa = 10.86STIPVLKK261 pKa = 9.73GVPVRR266 pKa = 11.84LTGSYY271 pKa = 7.75PTGSNTFEE279 pKa = 4.24SVSGSTAIIYY289 pKa = 9.86DD290 pKa = 3.85DD291 pKa = 3.67NALVV295 pKa = 3.35
MM1 pKa = 7.65ALKK4 pKa = 10.46QEE6 pKa = 4.1IFEE9 pKa = 4.34QNSTLFEE16 pKa = 4.1VLDD19 pKa = 4.11EE20 pKa = 4.47NEE22 pKa = 3.8VMEE25 pKa = 4.51IKK27 pKa = 10.3EE28 pKa = 4.12IQSSVTAVQTQVDD41 pKa = 4.02QQKK44 pKa = 10.33LQLDD48 pKa = 4.17GLARR52 pKa = 11.84RR53 pKa = 11.84VDD55 pKa = 3.92LNQARR60 pKa = 11.84NEE62 pKa = 4.02EE63 pKa = 4.13QFVNFNTMLIATNTEE78 pKa = 3.38VDD80 pKa = 3.5KK81 pKa = 11.63LKK83 pKa = 8.78TTTTTLSVSVNNLEE97 pKa = 4.06RR98 pKa = 11.84SVGEE102 pKa = 4.62LEE104 pKa = 4.22TTTQSNFDD112 pKa = 3.8TIFRR116 pKa = 11.84RR117 pKa = 11.84LAITEE122 pKa = 4.19EE123 pKa = 4.19TSSQNKK129 pKa = 9.92SEE131 pKa = 4.09IVTILDD137 pKa = 4.13KK138 pKa = 10.85IDD140 pKa = 4.31DD141 pKa = 4.41LEE143 pKa = 5.5HH144 pKa = 7.26SVEE147 pKa = 4.21SANDD151 pKa = 3.63EE152 pKa = 4.16IAEE155 pKa = 4.22LTDD158 pKa = 3.84SVEE161 pKa = 4.47DD162 pKa = 3.61NQTAITTLSGRR173 pKa = 11.84VTTLADD179 pKa = 3.11RR180 pKa = 11.84VTKK183 pKa = 10.78LEE185 pKa = 4.7NIVQSISPYY194 pKa = 8.79IQRR197 pKa = 11.84QTFYY201 pKa = 10.87RR202 pKa = 11.84IVYY205 pKa = 8.38YY206 pKa = 9.05KK207 pKa = 10.69TNGSMRR213 pKa = 11.84NVGFWANGYY222 pKa = 8.78VRR224 pKa = 11.84YY225 pKa = 10.65SMSFQVGTSATANFFTNLTITSVISSYY252 pKa = 11.14DD253 pKa = 3.35YY254 pKa = 10.86STIPVLKK261 pKa = 9.73GVPVRR266 pKa = 11.84LTGSYY271 pKa = 7.75PTGSNTFEE279 pKa = 4.24SVSGSTAIIYY289 pKa = 9.86DD290 pKa = 3.85DD291 pKa = 3.67NALVV295 pKa = 3.35
Molecular weight: 32.98 kDa
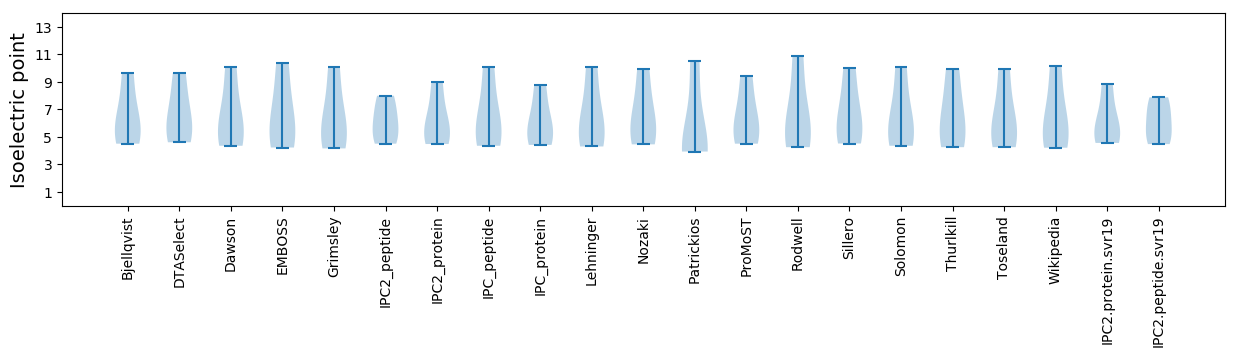
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075E230|A0A075E230_9VIRU VP1 OS=Drosophila immigrans Nora virus OX=1500866 GN=VP1 PE=4 SV=1
MM1 pKa = 7.04NQDD4 pKa = 3.19EE5 pKa = 4.48QKK7 pKa = 11.16KK8 pKa = 10.37KK9 pKa = 9.61GAQLKK14 pKa = 9.54RR15 pKa = 11.84VHH17 pKa = 5.76TVGTQVVDD25 pKa = 3.17TCNRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84QKK33 pKa = 8.27GTKK36 pKa = 8.6MEE38 pKa = 4.3VNFTVKK44 pKa = 10.27RR45 pKa = 11.84NNADD49 pKa = 3.42EE50 pKa = 4.12EE51 pKa = 4.56QKK53 pKa = 10.93FFVTEE58 pKa = 4.06RR59 pKa = 11.84VDD61 pKa = 3.95HH62 pKa = 6.46KK63 pKa = 11.24LEE65 pKa = 4.16KK66 pKa = 9.74QVKK69 pKa = 9.42AEE71 pKa = 3.91AKK73 pKa = 9.81QNHH76 pKa = 5.64TFIKK80 pKa = 10.11PKK82 pKa = 10.7LNFVDD87 pKa = 3.72KK88 pKa = 11.7VEE90 pKa = 4.06VEE92 pKa = 4.31KK93 pKa = 11.52VKK95 pKa = 9.65VTSRR99 pKa = 11.84GKK101 pKa = 9.87EE102 pKa = 3.4RR103 pKa = 11.84AANFAFFKK111 pKa = 10.86EE112 pKa = 4.29LVNSNKK118 pKa = 9.12VEE120 pKa = 4.18QKK122 pKa = 10.4WNVEE126 pKa = 4.01TEE128 pKa = 4.06KK129 pKa = 11.14EE130 pKa = 3.75IDD132 pKa = 3.66EE133 pKa = 4.31VDD135 pKa = 3.57LFFMKK140 pKa = 10.47KK141 pKa = 7.05KK142 pKa = 8.73TKK144 pKa = 9.54PFSGFSIKK152 pKa = 10.71SLGDD156 pKa = 3.52SLKK159 pKa = 10.76VMPDD163 pKa = 3.19EE164 pKa = 4.59KK165 pKa = 10.79HH166 pKa = 6.48ISPQTAMSSINEE178 pKa = 4.08NVATDD183 pKa = 3.59AVVDD187 pKa = 5.13VIALTAQLNQLIKK200 pKa = 10.44RR201 pKa = 11.84VEE203 pKa = 3.87ALEE206 pKa = 4.57LDD208 pKa = 3.68NKK210 pKa = 10.37QLVEE214 pKa = 4.23MNNQLRR220 pKa = 11.84QDD222 pKa = 3.58NEE224 pKa = 3.93KK225 pKa = 10.63LVNEE229 pKa = 5.72GITSIKK235 pKa = 10.03NDD237 pKa = 3.19KK238 pKa = 10.42KK239 pKa = 10.61EE240 pKa = 4.28KK241 pKa = 10.37KK242 pKa = 9.86NSEE245 pKa = 4.1NKK247 pKa = 9.75IEE249 pKa = 3.97QAISKK254 pKa = 7.45TASCMFKK261 pKa = 10.14IVAEE265 pKa = 4.5KK266 pKa = 10.73KK267 pKa = 10.0RR268 pKa = 11.84VTQVTPIVGEE278 pKa = 4.17EE279 pKa = 3.87QCLEE283 pKa = 4.02LQSTIKK289 pKa = 10.32RR290 pKa = 11.84KK291 pKa = 10.2YY292 pKa = 9.19KK293 pKa = 10.23EE294 pKa = 4.13KK295 pKa = 10.32PKK297 pKa = 10.73LPSVPSKK304 pKa = 10.8KK305 pKa = 9.54EE306 pKa = 3.68KK307 pKa = 10.6KK308 pKa = 10.26SIQQTKK314 pKa = 8.31IRR316 pKa = 11.84QWYY319 pKa = 8.62NFNKK323 pKa = 8.19QTIEE327 pKa = 3.88EE328 pKa = 4.23HH329 pKa = 5.39QKK331 pKa = 10.41EE332 pKa = 4.55VLSSVQTDD340 pKa = 3.18VTFAEE345 pKa = 4.59KK346 pKa = 10.6VKK348 pKa = 10.9EE349 pKa = 3.88NGIPKK354 pKa = 9.89QKK356 pKa = 10.41LRR358 pKa = 11.84YY359 pKa = 6.93TVSTTVEE366 pKa = 4.08KK367 pKa = 10.69QPKK370 pKa = 9.66KK371 pKa = 10.43SIHH374 pKa = 6.22HH375 pKa = 6.04YY376 pKa = 8.86GYY378 pKa = 10.84KK379 pKa = 9.72PAGVPNKK386 pKa = 9.75IWWKK390 pKa = 9.09WVTTGTAAEE399 pKa = 4.56AYY401 pKa = 9.42KK402 pKa = 10.48KK403 pKa = 10.76AQTFLFKK410 pKa = 10.08MFKK413 pKa = 10.32RR414 pKa = 11.84EE415 pKa = 4.07MIVFRR420 pKa = 11.84QKK422 pKa = 10.0WICHH426 pKa = 4.84TKK428 pKa = 10.22EE429 pKa = 3.7FNPYY433 pKa = 9.49LSEE436 pKa = 4.24PKK438 pKa = 9.76MVWLEE443 pKa = 3.89NTNEE447 pKa = 3.74YY448 pKa = 10.2PYY450 pKa = 11.08DD451 pKa = 3.8VNVLFEE457 pKa = 5.13FIKK460 pKa = 9.63KK461 pKa = 8.36WRR463 pKa = 11.84MLVTSYY469 pKa = 11.35KK470 pKa = 9.44PGKK473 pKa = 9.4PITNDD478 pKa = 2.5WYY480 pKa = 11.13KK481 pKa = 11.26NKK483 pKa = 10.17QNN485 pKa = 3.82
MM1 pKa = 7.04NQDD4 pKa = 3.19EE5 pKa = 4.48QKK7 pKa = 11.16KK8 pKa = 10.37KK9 pKa = 9.61GAQLKK14 pKa = 9.54RR15 pKa = 11.84VHH17 pKa = 5.76TVGTQVVDD25 pKa = 3.17TCNRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84QKK33 pKa = 8.27GTKK36 pKa = 8.6MEE38 pKa = 4.3VNFTVKK44 pKa = 10.27RR45 pKa = 11.84NNADD49 pKa = 3.42EE50 pKa = 4.12EE51 pKa = 4.56QKK53 pKa = 10.93FFVTEE58 pKa = 4.06RR59 pKa = 11.84VDD61 pKa = 3.95HH62 pKa = 6.46KK63 pKa = 11.24LEE65 pKa = 4.16KK66 pKa = 9.74QVKK69 pKa = 9.42AEE71 pKa = 3.91AKK73 pKa = 9.81QNHH76 pKa = 5.64TFIKK80 pKa = 10.11PKK82 pKa = 10.7LNFVDD87 pKa = 3.72KK88 pKa = 11.7VEE90 pKa = 4.06VEE92 pKa = 4.31KK93 pKa = 11.52VKK95 pKa = 9.65VTSRR99 pKa = 11.84GKK101 pKa = 9.87EE102 pKa = 3.4RR103 pKa = 11.84AANFAFFKK111 pKa = 10.86EE112 pKa = 4.29LVNSNKK118 pKa = 9.12VEE120 pKa = 4.18QKK122 pKa = 10.4WNVEE126 pKa = 4.01TEE128 pKa = 4.06KK129 pKa = 11.14EE130 pKa = 3.75IDD132 pKa = 3.66EE133 pKa = 4.31VDD135 pKa = 3.57LFFMKK140 pKa = 10.47KK141 pKa = 7.05KK142 pKa = 8.73TKK144 pKa = 9.54PFSGFSIKK152 pKa = 10.71SLGDD156 pKa = 3.52SLKK159 pKa = 10.76VMPDD163 pKa = 3.19EE164 pKa = 4.59KK165 pKa = 10.79HH166 pKa = 6.48ISPQTAMSSINEE178 pKa = 4.08NVATDD183 pKa = 3.59AVVDD187 pKa = 5.13VIALTAQLNQLIKK200 pKa = 10.44RR201 pKa = 11.84VEE203 pKa = 3.87ALEE206 pKa = 4.57LDD208 pKa = 3.68NKK210 pKa = 10.37QLVEE214 pKa = 4.23MNNQLRR220 pKa = 11.84QDD222 pKa = 3.58NEE224 pKa = 3.93KK225 pKa = 10.63LVNEE229 pKa = 5.72GITSIKK235 pKa = 10.03NDD237 pKa = 3.19KK238 pKa = 10.42KK239 pKa = 10.61EE240 pKa = 4.28KK241 pKa = 10.37KK242 pKa = 9.86NSEE245 pKa = 4.1NKK247 pKa = 9.75IEE249 pKa = 3.97QAISKK254 pKa = 7.45TASCMFKK261 pKa = 10.14IVAEE265 pKa = 4.5KK266 pKa = 10.73KK267 pKa = 10.0RR268 pKa = 11.84VTQVTPIVGEE278 pKa = 4.17EE279 pKa = 3.87QCLEE283 pKa = 4.02LQSTIKK289 pKa = 10.32RR290 pKa = 11.84KK291 pKa = 10.2YY292 pKa = 9.19KK293 pKa = 10.23EE294 pKa = 4.13KK295 pKa = 10.32PKK297 pKa = 10.73LPSVPSKK304 pKa = 10.8KK305 pKa = 9.54EE306 pKa = 3.68KK307 pKa = 10.6KK308 pKa = 10.26SIQQTKK314 pKa = 8.31IRR316 pKa = 11.84QWYY319 pKa = 8.62NFNKK323 pKa = 8.19QTIEE327 pKa = 3.88EE328 pKa = 4.23HH329 pKa = 5.39QKK331 pKa = 10.41EE332 pKa = 4.55VLSSVQTDD340 pKa = 3.18VTFAEE345 pKa = 4.59KK346 pKa = 10.6VKK348 pKa = 10.9EE349 pKa = 3.88NGIPKK354 pKa = 9.89QKK356 pKa = 10.41LRR358 pKa = 11.84YY359 pKa = 6.93TVSTTVEE366 pKa = 4.08KK367 pKa = 10.69QPKK370 pKa = 9.66KK371 pKa = 10.43SIHH374 pKa = 6.22HH375 pKa = 6.04YY376 pKa = 8.86GYY378 pKa = 10.84KK379 pKa = 9.72PAGVPNKK386 pKa = 9.75IWWKK390 pKa = 9.09WVTTGTAAEE399 pKa = 4.56AYY401 pKa = 9.42KK402 pKa = 10.48KK403 pKa = 10.76AQTFLFKK410 pKa = 10.08MFKK413 pKa = 10.32RR414 pKa = 11.84EE415 pKa = 4.07MIVFRR420 pKa = 11.84QKK422 pKa = 10.0WICHH426 pKa = 4.84TKK428 pKa = 10.22EE429 pKa = 3.7FNPYY433 pKa = 9.49LSEE436 pKa = 4.24PKK438 pKa = 9.76MVWLEE443 pKa = 3.89NTNEE447 pKa = 3.74YY448 pKa = 10.2PYY450 pKa = 11.08DD451 pKa = 3.8VNVLFEE457 pKa = 5.13FIKK460 pKa = 9.63KK461 pKa = 8.36WRR463 pKa = 11.84MLVTSYY469 pKa = 11.35KK470 pKa = 9.44PGKK473 pKa = 9.4PITNDD478 pKa = 2.5WYY480 pKa = 11.13KK481 pKa = 11.26NKK483 pKa = 10.17QNN485 pKa = 3.82
Molecular weight: 56.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3761 |
295 |
2043 |
940.3 |
106.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.275 ± 0.634 | 1.09 ± 0.264 |
4.892 ± 0.395 | 7.392 ± 0.431 |
4.201 ± 0.15 | 5.238 ± 0.677 |
1.462 ± 0.216 | 6.7 ± 0.442 |
7.471 ± 1.956 | 8.349 ± 0.85 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.994 ± 0.071 | 5.477 ± 0.48 |
4.121 ± 1.139 | 4.866 ± 0.611 |
4.307 ± 0.175 | 6.036 ± 0.659 |
7.312 ± 0.98 | 7.844 ± 0.522 |
1.462 ± 0.253 | 3.51 ± 0.456 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
