
Posidoniimonas polymericola
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Lacipirellulaceae; Posidoniimonas
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
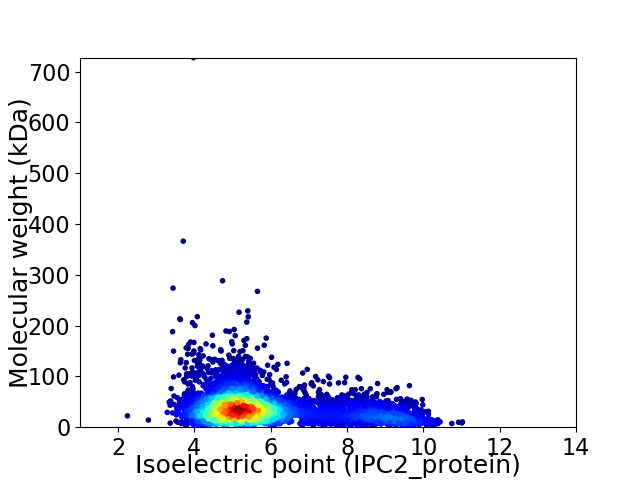
Virtual 2D-PAGE plot for 4870 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
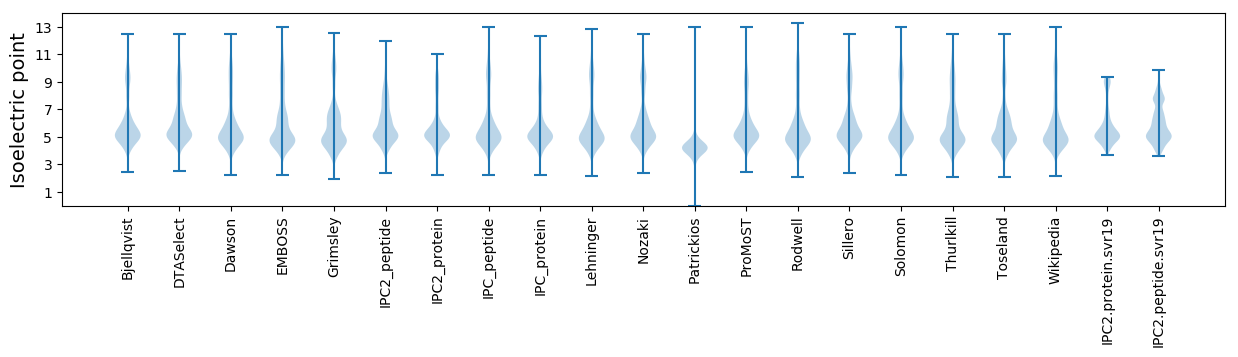
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C5YKX3|A0A5C5YKX3_9BACT Putative acetyltransferase OS=Posidoniimonas polymericola OX=2528002 GN=Pla123a_29780 PE=4 SV=1
MM1 pKa = 7.38CRR3 pKa = 11.84SIAAITLATLLASQGVCHH21 pKa = 6.06AVVVASLEE29 pKa = 4.49TNVAPAGTAGDD40 pKa = 4.24PYY42 pKa = 11.34NPTFTAGGPSTTDD55 pKa = 3.41LLEE58 pKa = 4.54GLLPTVAIGNLTQEE72 pKa = 4.19NSLGASALTNGTVATAYY89 pKa = 10.48GSGDD93 pKa = 3.52ANSAHH98 pKa = 6.99IAYY101 pKa = 7.55ATGGPGEE108 pKa = 4.36EE109 pKa = 3.66VTYY112 pKa = 10.97ALGGIYY118 pKa = 10.35DD119 pKa = 4.69LDD121 pKa = 4.45SIVIFGGWNDD131 pKa = 3.68AGRR134 pKa = 11.84DD135 pKa = 3.64AQSYY139 pKa = 10.07DD140 pKa = 4.05LYY142 pKa = 11.04VSSDD146 pKa = 3.07GAQNFSLLTNYY157 pKa = 10.61NNGNGEE163 pKa = 4.1EE164 pKa = 4.17QGVVTTPVSHH174 pKa = 6.41RR175 pKa = 11.84VEE177 pKa = 4.25FTEE180 pKa = 5.88DD181 pKa = 3.35SLPNLATGVTHH192 pKa = 6.63VKK194 pKa = 10.32VDD196 pKa = 4.0FLDD199 pKa = 3.66VEE201 pKa = 4.28NGYY204 pKa = 8.04TGYY207 pKa = 9.28TEE209 pKa = 4.28IDD211 pKa = 3.56VFGSLVNVPGDD222 pKa = 3.54ADD224 pKa = 3.75GDD226 pKa = 4.13GDD228 pKa = 4.3VDD230 pKa = 4.69LDD232 pKa = 4.02DD233 pKa = 5.52FFVISNHH240 pKa = 5.89FFTTPSAIGLDD251 pKa = 3.74GDD253 pKa = 4.12VEE255 pKa = 4.27PDD257 pKa = 3.54NIVDD261 pKa = 3.79EE262 pKa = 4.62KK263 pKa = 11.4DD264 pKa = 3.1FRR266 pKa = 11.84LWKK269 pKa = 10.01SVASASVLAEE279 pKa = 4.21FAALGVPEE287 pKa = 5.15PSTLALIGVSMGLLAGRR304 pKa = 11.84RR305 pKa = 11.84RR306 pKa = 11.84GG307 pKa = 3.38
MM1 pKa = 7.38CRR3 pKa = 11.84SIAAITLATLLASQGVCHH21 pKa = 6.06AVVVASLEE29 pKa = 4.49TNVAPAGTAGDD40 pKa = 4.24PYY42 pKa = 11.34NPTFTAGGPSTTDD55 pKa = 3.41LLEE58 pKa = 4.54GLLPTVAIGNLTQEE72 pKa = 4.19NSLGASALTNGTVATAYY89 pKa = 10.48GSGDD93 pKa = 3.52ANSAHH98 pKa = 6.99IAYY101 pKa = 7.55ATGGPGEE108 pKa = 4.36EE109 pKa = 3.66VTYY112 pKa = 10.97ALGGIYY118 pKa = 10.35DD119 pKa = 4.69LDD121 pKa = 4.45SIVIFGGWNDD131 pKa = 3.68AGRR134 pKa = 11.84DD135 pKa = 3.64AQSYY139 pKa = 10.07DD140 pKa = 4.05LYY142 pKa = 11.04VSSDD146 pKa = 3.07GAQNFSLLTNYY157 pKa = 10.61NNGNGEE163 pKa = 4.1EE164 pKa = 4.17QGVVTTPVSHH174 pKa = 6.41RR175 pKa = 11.84VEE177 pKa = 4.25FTEE180 pKa = 5.88DD181 pKa = 3.35SLPNLATGVTHH192 pKa = 6.63VKK194 pKa = 10.32VDD196 pKa = 4.0FLDD199 pKa = 3.66VEE201 pKa = 4.28NGYY204 pKa = 8.04TGYY207 pKa = 9.28TEE209 pKa = 4.28IDD211 pKa = 3.56VFGSLVNVPGDD222 pKa = 3.54ADD224 pKa = 3.75GDD226 pKa = 4.13GDD228 pKa = 4.3VDD230 pKa = 4.69LDD232 pKa = 4.02DD233 pKa = 5.52FFVISNHH240 pKa = 5.89FFTTPSAIGLDD251 pKa = 3.74GDD253 pKa = 4.12VEE255 pKa = 4.27PDD257 pKa = 3.54NIVDD261 pKa = 3.79EE262 pKa = 4.62KK263 pKa = 11.4DD264 pKa = 3.1FRR266 pKa = 11.84LWKK269 pKa = 10.01SVASASVLAEE279 pKa = 4.21FAALGVPEE287 pKa = 5.15PSTLALIGVSMGLLAGRR304 pKa = 11.84RR305 pKa = 11.84RR306 pKa = 11.84GG307 pKa = 3.38
Molecular weight: 31.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C5YSW6|A0A5C5YSW6_9BACT Quinolinate synthase A OS=Posidoniimonas polymericola OX=2528002 GN=nadA PE=3 SV=1
MM1 pKa = 7.07NRR3 pKa = 11.84KK4 pKa = 8.76NLVVAAVLASAFALGMLTSKK24 pKa = 10.28QASAGKK30 pKa = 9.95YY31 pKa = 9.49GPAGLGANLHH41 pKa = 6.26RR42 pKa = 11.84RR43 pKa = 11.84HH44 pKa = 6.14VMKK47 pKa = 10.59QATKK51 pKa = 9.23NARR54 pKa = 11.84RR55 pKa = 11.84GLPVRR60 pKa = 11.84KK61 pKa = 9.33SAIVWLSAPQSNLRR75 pKa = 11.84WARR78 pKa = 3.38
MM1 pKa = 7.07NRR3 pKa = 11.84KK4 pKa = 8.76NLVVAAVLASAFALGMLTSKK24 pKa = 10.28QASAGKK30 pKa = 9.95YY31 pKa = 9.49GPAGLGANLHH41 pKa = 6.26RR42 pKa = 11.84RR43 pKa = 11.84HH44 pKa = 6.14VMKK47 pKa = 10.59QATKK51 pKa = 9.23NARR54 pKa = 11.84RR55 pKa = 11.84GLPVRR60 pKa = 11.84KK61 pKa = 9.33SAIVWLSAPQSNLRR75 pKa = 11.84WARR78 pKa = 3.38
Molecular weight: 8.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1793801 |
30 |
6968 |
368.3 |
40.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.531 ± 0.041 | 1.092 ± 0.014 |
6.292 ± 0.029 | 6.16 ± 0.03 |
3.367 ± 0.019 | 8.628 ± 0.043 |
2.044 ± 0.017 | 4.036 ± 0.022 |
2.84 ± 0.028 | 10.031 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.987 ± 0.015 | 2.892 ± 0.023 |
5.582 ± 0.032 | 3.794 ± 0.023 |
6.746 ± 0.038 | 6.052 ± 0.029 |
5.352 ± 0.029 | 7.519 ± 0.026 |
1.538 ± 0.014 | 2.519 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
