
Cyanobacterium aponinum (strain PCC 10605)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Chroococcales; Geminocystaceae; Cyanobacterium; Cyanobacterium aponinum
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
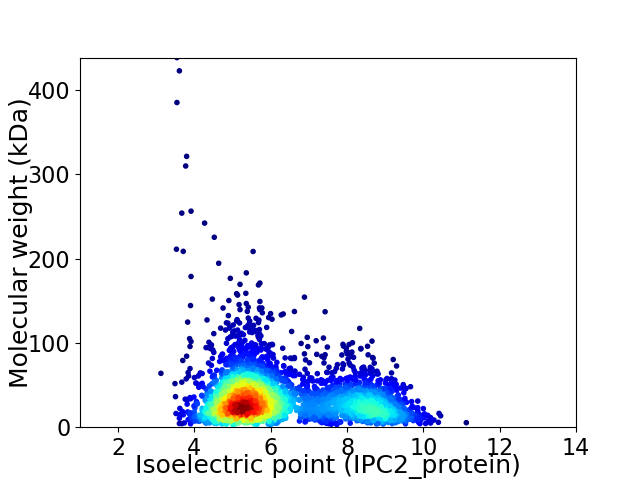
Virtual 2D-PAGE plot for 3415 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
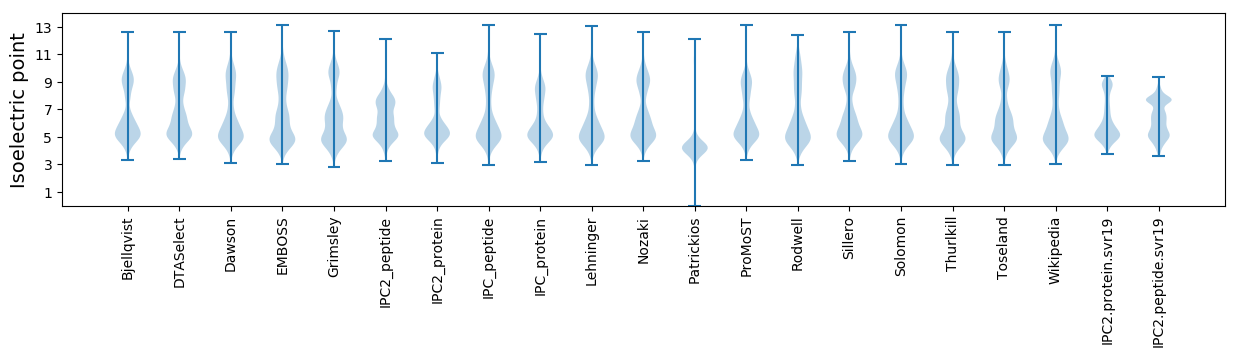
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9Z106|K9Z106_CYAAP Uncharacterized protein OS=Cyanobacterium aponinum (strain PCC 10605) OX=755178 GN=Cyan10605_0267 PE=4 SV=1
MM1 pKa = 7.33SLVTGDD7 pKa = 3.31IDD9 pKa = 4.48GDD11 pKa = 4.21GIEE14 pKa = 5.0DD15 pKa = 4.05LVIGAPNVNNGDD27 pKa = 3.69GIVYY31 pKa = 8.51VIRR34 pKa = 11.84GSYY37 pKa = 9.5IADD40 pKa = 3.37NQGQIININSDD51 pKa = 2.87NSFSDD56 pKa = 3.57QKK58 pKa = 10.31NTASNNIGFVFNPTVTGAYY77 pKa = 9.13FGYY80 pKa = 10.67AVAVGNFDD88 pKa = 4.12GNGSLDD94 pKa = 3.87LAISSPGASKK104 pKa = 11.25GDD106 pKa = 3.42GLVYY110 pKa = 10.08IAHH113 pKa = 6.6NNSEE117 pKa = 4.44LQTFATGSNGEE128 pKa = 3.99NLGYY132 pKa = 10.61SLAVSQANGKK142 pKa = 9.28QSFSGNTTIDD152 pKa = 3.72DD153 pKa = 5.26LIVGAPSYY161 pKa = 10.97ASSVSNQWVGADD173 pKa = 3.48QLPSEE178 pKa = 4.42NQNLFPNTTSTAIGKK193 pKa = 9.8VYY195 pKa = 10.84VFGNNQTTPLYY206 pKa = 10.3SFTGSILPSTNGTAEE221 pKa = 3.81NSFTGSALASDD232 pKa = 4.53DD233 pKa = 3.65WNLDD237 pKa = 3.16GARR240 pKa = 11.84DD241 pKa = 3.78LAISAPGSDD250 pKa = 3.39NSDD253 pKa = 3.06GLVYY257 pKa = 10.21VVKK260 pKa = 10.56GGKK263 pKa = 7.04PTSGDD268 pKa = 3.3LDD270 pKa = 4.31SISNLIILGGLPFSKK285 pKa = 9.43TGSEE289 pKa = 3.84IASAGDD295 pKa = 3.52VNGDD299 pKa = 3.29GYY301 pKa = 11.62EE302 pKa = 4.23DD303 pKa = 4.15FLVGAPQGLMGVGQSYY319 pKa = 11.14LLFGPLDD326 pKa = 4.36LDD328 pKa = 3.93SQGTLFNLNVTATDD342 pKa = 3.45SKK344 pKa = 11.0KK345 pKa = 9.05TFLLNGSQPYY355 pKa = 10.05QLTGSALSGIGDD367 pKa = 3.82VNNDD371 pKa = 3.23NVDD374 pKa = 3.81DD375 pKa = 5.0LMITAPNAQQLYY387 pKa = 10.75AVFGHH392 pKa = 5.97QWLADD397 pKa = 3.95DD398 pKa = 5.52GSIKK402 pKa = 10.61LADD405 pKa = 3.3ISADD409 pKa = 3.16NGFVIDD415 pKa = 3.92GDD417 pKa = 4.1LYY419 pKa = 10.83KK420 pKa = 11.12ANSKK424 pKa = 10.21SLNGNGNNVLILGDD438 pKa = 3.58INGDD442 pKa = 3.31GFADD446 pKa = 3.76VLSGGSEE453 pKa = 4.07DD454 pKa = 3.61GAVIIFGSSTEE465 pKa = 4.01NLLDD469 pKa = 3.99ASVGSNDD476 pKa = 5.62LILSVANHH484 pKa = 6.9SIQDD488 pKa = 3.69FASLGDD494 pKa = 3.79YY495 pKa = 11.05NGDD498 pKa = 3.53GLQDD502 pKa = 3.76FGVIDD507 pKa = 5.09DD508 pKa = 5.15DD509 pKa = 4.55NNFYY513 pKa = 10.56IQLGSNNLNSLGNLSLSNPSVTSIIEE539 pKa = 4.48AIEE542 pKa = 4.17LGDD545 pKa = 3.94YY546 pKa = 11.18NGDD549 pKa = 3.68GYY551 pKa = 11.41DD552 pKa = 5.27DD553 pKa = 3.9ILLNTSSNSRR563 pKa = 11.84IYY565 pKa = 10.75LGNEE569 pKa = 3.58EE570 pKa = 4.8GNVNNFLTFNPVGGNSFSDD589 pKa = 3.68YY590 pKa = 10.38TFGSIGDD597 pKa = 3.76IDD599 pKa = 4.27GDD601 pKa = 4.38GYY603 pKa = 11.33QDD605 pKa = 3.21IATGLPQSNFTIQDD619 pKa = 3.29AANGQFTIYY628 pKa = 9.92TNRR631 pKa = 11.84GNQSILPPVTPSLSNFPSNDD651 pKa = 3.5YY652 pKa = 11.1NPTIASYY659 pKa = 10.52GVPSQQSPVPPNFAVYY675 pKa = 9.87QGYY678 pKa = 10.43LYY680 pKa = 7.91MTFKK684 pKa = 11.35GNGNDD689 pKa = 4.67DD690 pKa = 3.86INVMRR695 pKa = 11.84SRR697 pKa = 11.84DD698 pKa = 3.51GNTWEE703 pKa = 4.12NQVVLTSLQTPYY715 pKa = 10.92SPSLVVFEE723 pKa = 5.04DD724 pKa = 3.28KK725 pKa = 10.94LYY727 pKa = 10.81LIHH730 pKa = 6.63TFEE733 pKa = 5.87DD734 pKa = 3.36STIYY738 pKa = 9.1ITPFTGDD745 pKa = 3.76DD746 pKa = 3.35NSISFGEE753 pKa = 4.3YY754 pKa = 8.03TALIADD760 pKa = 5.04AGTSPTPIVFNNKK773 pKa = 9.63LYY775 pKa = 10.47IFYY778 pKa = 10.36IADD781 pKa = 3.62GDD783 pKa = 3.98DD784 pKa = 3.46QINYY788 pKa = 7.19ITLSDD793 pKa = 3.91PANVTEE799 pKa = 4.84FGYY802 pKa = 11.08DD803 pKa = 2.99EE804 pKa = 4.35FNYY807 pKa = 10.12INGQQSFDD815 pKa = 3.37RR816 pKa = 11.84VGATLTPDD824 pKa = 3.49GEE826 pKa = 4.57KK827 pKa = 10.73LVLAYY832 pKa = 9.88KK833 pKa = 10.46NSNKK837 pKa = 9.99ADD839 pKa = 3.23IYY841 pKa = 9.79ITNFDD846 pKa = 3.52GSNWSEE852 pKa = 3.76GKK854 pKa = 9.71IVQDD858 pKa = 3.46SQNNNNTIEE867 pKa = 4.29TTDD870 pKa = 3.74GVSLTTVGNNIYY882 pKa = 10.26LAYY885 pKa = 10.11EE886 pKa = 4.13GTKK889 pKa = 10.69PNDD892 pKa = 2.95LRR894 pKa = 11.84YY895 pKa = 9.69IYY897 pKa = 10.72SQDD900 pKa = 4.24SGNSWEE906 pKa = 4.44NDD908 pKa = 2.82RR909 pKa = 11.84NTPNQLVNGGASLTFFQSSLYY930 pKa = 10.22FGYY933 pKa = 10.44SSQVTSNAPLYY944 pKa = 10.41VSYY947 pKa = 10.96SEE949 pKa = 5.27PIYY952 pKa = 10.02NTNQTQQLGEE962 pKa = 3.92QLYY965 pKa = 11.05SIGDD969 pKa = 3.65FNGDD973 pKa = 3.67GIEE976 pKa = 4.53DD977 pKa = 3.89FAIVAQGYY985 pKa = 9.29IADD988 pKa = 4.75LGIIDD993 pKa = 5.37LSAGLQIRR1001 pKa = 11.84NNQGAILIYY1010 pKa = 10.14YY1011 pKa = 9.31GNANGISNSAKK1022 pKa = 9.64PDD1024 pKa = 3.5VVLVNPPVTDD1034 pKa = 3.73NQSLLYY1040 pKa = 9.97EE1041 pKa = 4.41INRR1044 pKa = 11.84FTAIGDD1050 pKa = 3.82INGDD1054 pKa = 3.47GYY1056 pKa = 11.49DD1057 pKa = 3.94DD1058 pKa = 4.16VAVSSPTTKK1067 pKa = 10.6SNEE1070 pKa = 3.35GSIFVVFGGSSWQSTYY1086 pKa = 11.3SATNPFDD1093 pKa = 5.04LSSLSNNQSNSTTDD1107 pKa = 3.16NSNTNGFLITGLPASQAGISLSGGEE1132 pKa = 4.65DD1133 pKa = 3.46VNGDD1137 pKa = 3.49GFDD1140 pKa = 4.23DD1141 pKa = 4.16FTVGAPGNDD1150 pKa = 3.62DD1151 pKa = 3.83DD1152 pKa = 4.23LTYY1155 pKa = 11.17VIFGSDD1161 pKa = 2.95FTQQVNQTGTIGRR1174 pKa = 11.84DD1175 pKa = 3.15RR1176 pKa = 11.84MIGTPTGEE1184 pKa = 3.63IFIAGEE1190 pKa = 4.16GNDD1193 pKa = 3.74RR1194 pKa = 11.84IYY1196 pKa = 11.46SNGGVDD1202 pKa = 3.4VVYY1205 pKa = 10.93ASVGNDD1211 pKa = 2.95FVTVNDD1217 pKa = 3.87TYY1219 pKa = 11.02FRR1221 pKa = 11.84RR1222 pKa = 11.84LDD1224 pKa = 3.66GGTGIDD1230 pKa = 3.33TLKK1233 pKa = 9.53FTGYY1237 pKa = 10.5NGQNWDD1243 pKa = 4.22LTTLSPGNRR1252 pKa = 11.84LRR1254 pKa = 11.84NFEE1257 pKa = 4.02ILDD1260 pKa = 3.43IKK1262 pKa = 11.13DD1263 pKa = 3.49YY1264 pKa = 11.74GEE1266 pKa = 4.01NTLTLNSLTVTQLSSNNTITVLMDD1290 pKa = 4.47KK1291 pKa = 10.71EE1292 pKa = 4.2DD1293 pKa = 4.0TLNLSADD1300 pKa = 3.86FVNEE1304 pKa = 4.45GIINRR1309 pKa = 11.84NGEE1312 pKa = 4.21TYY1314 pKa = 9.69QKK1316 pKa = 10.91YY1317 pKa = 9.98RR1318 pKa = 11.84IDD1320 pKa = 3.67GATVLILEE1328 pKa = 5.73DD1329 pKa = 3.39IVQTKK1334 pKa = 10.76AFTPLSLSLYY1344 pKa = 8.35NTGVDD1349 pKa = 4.88DD1350 pKa = 6.19SGTPLNNNSTGDD1362 pKa = 3.41PHH1364 pKa = 8.21YY1365 pKa = 10.11EE1366 pKa = 3.92LVSAPDD1372 pKa = 3.95GSSSNILVTNSGDD1385 pKa = 4.16GISGWISPDD1394 pKa = 3.18NNNSDD1399 pKa = 3.6SSPPGDD1405 pKa = 3.39YY1406 pKa = 10.2TYY1408 pKa = 10.19RR1409 pKa = 11.84TTFNLPDD1416 pKa = 3.97TVNLSQFFIDD1426 pKa = 5.46GEE1428 pKa = 4.2WHH1430 pKa = 6.4TDD1432 pKa = 3.41DD1433 pKa = 4.38EE1434 pKa = 5.76GITITLNGIEE1444 pKa = 4.85QYY1446 pKa = 10.73LASNPNFNTDD1456 pKa = 3.91FVPFTINEE1464 pKa = 3.95GFIAGEE1470 pKa = 4.24NILEE1474 pKa = 4.13FTVNNAGTEE1483 pKa = 4.0INPTVLRR1490 pKa = 11.84VKK1492 pKa = 10.47FDD1494 pKa = 3.41NAFDD1498 pKa = 4.39SNIQFSAPSTNIPQAILNPEE1518 pKa = 4.04NVSQVSSANNSVVSIDD1534 pKa = 3.4EE1535 pKa = 4.43SNNITEE1541 pKa = 4.32VSSSLFANTSNNEE1554 pKa = 3.87DD1555 pKa = 3.65STTNLYY1561 pKa = 7.72VTNPTVNEE1569 pKa = 3.67ADD1571 pKa = 3.85GEE1573 pKa = 4.84VKK1575 pKa = 9.33FTIQRR1580 pKa = 11.84TGNLDD1585 pKa = 3.38KK1586 pKa = 10.89YY1587 pKa = 9.41VQIHH1591 pKa = 5.85YY1592 pKa = 7.6VTQDD1596 pKa = 2.69GRR1598 pKa = 11.84AKK1600 pKa = 10.57AGNDD1604 pKa = 3.16YY1605 pKa = 10.93HH1606 pKa = 6.17PTVGKK1611 pKa = 10.51AIFTPGEE1618 pKa = 3.75SSINVSVPLILDD1630 pKa = 3.55DD1631 pKa = 4.75VYY1633 pKa = 11.37TGTKK1637 pKa = 10.25DD1638 pKa = 2.64IGLFVTLEE1646 pKa = 4.36KK1647 pKa = 10.59EE1648 pKa = 4.12SHH1650 pKa = 6.04TPLIDD1655 pKa = 3.36EE1656 pKa = 4.92FNIHH1660 pKa = 7.06ADD1662 pKa = 3.65FNNGIINSWHH1672 pKa = 6.8HH1673 pKa = 5.51IQEE1676 pKa = 4.49EE1677 pKa = 4.72STHH1680 pKa = 6.34PLSLINGEE1688 pKa = 4.03LEE1690 pKa = 4.12FRR1692 pKa = 11.84VTATEE1697 pKa = 4.36GEE1699 pKa = 4.49AEE1701 pKa = 4.22VKK1703 pKa = 10.53LYY1705 pKa = 10.83FDD1707 pKa = 4.84GNHH1710 pKa = 5.83EE1711 pKa = 4.29FNRR1714 pKa = 11.84YY1715 pKa = 9.86YY1716 pKa = 10.29IFNTQNQRR1724 pKa = 11.84YY1725 pKa = 7.95EE1726 pKa = 4.02VFDD1729 pKa = 4.35FNGDD1733 pKa = 3.11TGAEE1737 pKa = 4.21LFDD1740 pKa = 4.89EE1741 pKa = 5.92DD1742 pKa = 5.74DD1743 pKa = 3.96DD1744 pKa = 5.16SNYY1747 pKa = 10.8EE1748 pKa = 4.01GVILHH1753 pKa = 6.2LQDD1756 pKa = 4.48GSEE1759 pKa = 4.17HH1760 pKa = 6.9DD1761 pKa = 4.37LDD1763 pKa = 4.04GARR1766 pKa = 11.84NGVIIKK1772 pKa = 10.28RR1773 pKa = 11.84GFFAGGNPPEE1783 pKa = 4.31TVLNKK1788 pKa = 9.79PMYY1791 pKa = 10.18RR1792 pKa = 11.84FRR1794 pKa = 11.84NSDD1797 pKa = 3.61YY1798 pKa = 10.75DD1799 pKa = 3.54TGAYY1803 pKa = 10.07LYY1805 pKa = 10.44AGEE1808 pKa = 4.86IEE1810 pKa = 4.33SEE1812 pKa = 4.64SIRR1815 pKa = 11.84EE1816 pKa = 3.89NYY1818 pKa = 9.48PNFVEE1823 pKa = 4.5EE1824 pKa = 4.73GFAFYY1829 pKa = 11.16VSTTPQDD1836 pKa = 3.42GLITFNRR1843 pKa = 11.84FQNLDD1848 pKa = 3.41YY1849 pKa = 10.76PGSYY1853 pKa = 10.3LYY1855 pKa = 10.87AEE1857 pKa = 4.38EE1858 pKa = 4.72TEE1860 pKa = 4.35SEE1862 pKa = 4.74SIRR1865 pKa = 11.84EE1866 pKa = 3.94NYY1868 pKa = 9.48PNFVEE1873 pKa = 4.42EE1874 pKa = 5.28GIAFYY1879 pKa = 10.18AYY1881 pKa = 9.46PQGSQMADD1889 pKa = 3.29VISRR1893 pKa = 11.84FQSNDD1898 pKa = 2.94LLGSYY1903 pKa = 10.14LYY1905 pKa = 10.25TSEE1908 pKa = 5.1PEE1910 pKa = 3.8TDD1912 pKa = 3.5NVISNYY1918 pKa = 10.84ANFNLEE1924 pKa = 4.17GIAFEE1929 pKa = 4.58AVVAA1933 pKa = 4.42
MM1 pKa = 7.33SLVTGDD7 pKa = 3.31IDD9 pKa = 4.48GDD11 pKa = 4.21GIEE14 pKa = 5.0DD15 pKa = 4.05LVIGAPNVNNGDD27 pKa = 3.69GIVYY31 pKa = 8.51VIRR34 pKa = 11.84GSYY37 pKa = 9.5IADD40 pKa = 3.37NQGQIININSDD51 pKa = 2.87NSFSDD56 pKa = 3.57QKK58 pKa = 10.31NTASNNIGFVFNPTVTGAYY77 pKa = 9.13FGYY80 pKa = 10.67AVAVGNFDD88 pKa = 4.12GNGSLDD94 pKa = 3.87LAISSPGASKK104 pKa = 11.25GDD106 pKa = 3.42GLVYY110 pKa = 10.08IAHH113 pKa = 6.6NNSEE117 pKa = 4.44LQTFATGSNGEE128 pKa = 3.99NLGYY132 pKa = 10.61SLAVSQANGKK142 pKa = 9.28QSFSGNTTIDD152 pKa = 3.72DD153 pKa = 5.26LIVGAPSYY161 pKa = 10.97ASSVSNQWVGADD173 pKa = 3.48QLPSEE178 pKa = 4.42NQNLFPNTTSTAIGKK193 pKa = 9.8VYY195 pKa = 10.84VFGNNQTTPLYY206 pKa = 10.3SFTGSILPSTNGTAEE221 pKa = 3.81NSFTGSALASDD232 pKa = 4.53DD233 pKa = 3.65WNLDD237 pKa = 3.16GARR240 pKa = 11.84DD241 pKa = 3.78LAISAPGSDD250 pKa = 3.39NSDD253 pKa = 3.06GLVYY257 pKa = 10.21VVKK260 pKa = 10.56GGKK263 pKa = 7.04PTSGDD268 pKa = 3.3LDD270 pKa = 4.31SISNLIILGGLPFSKK285 pKa = 9.43TGSEE289 pKa = 3.84IASAGDD295 pKa = 3.52VNGDD299 pKa = 3.29GYY301 pKa = 11.62EE302 pKa = 4.23DD303 pKa = 4.15FLVGAPQGLMGVGQSYY319 pKa = 11.14LLFGPLDD326 pKa = 4.36LDD328 pKa = 3.93SQGTLFNLNVTATDD342 pKa = 3.45SKK344 pKa = 11.0KK345 pKa = 9.05TFLLNGSQPYY355 pKa = 10.05QLTGSALSGIGDD367 pKa = 3.82VNNDD371 pKa = 3.23NVDD374 pKa = 3.81DD375 pKa = 5.0LMITAPNAQQLYY387 pKa = 10.75AVFGHH392 pKa = 5.97QWLADD397 pKa = 3.95DD398 pKa = 5.52GSIKK402 pKa = 10.61LADD405 pKa = 3.3ISADD409 pKa = 3.16NGFVIDD415 pKa = 3.92GDD417 pKa = 4.1LYY419 pKa = 10.83KK420 pKa = 11.12ANSKK424 pKa = 10.21SLNGNGNNVLILGDD438 pKa = 3.58INGDD442 pKa = 3.31GFADD446 pKa = 3.76VLSGGSEE453 pKa = 4.07DD454 pKa = 3.61GAVIIFGSSTEE465 pKa = 4.01NLLDD469 pKa = 3.99ASVGSNDD476 pKa = 5.62LILSVANHH484 pKa = 6.9SIQDD488 pKa = 3.69FASLGDD494 pKa = 3.79YY495 pKa = 11.05NGDD498 pKa = 3.53GLQDD502 pKa = 3.76FGVIDD507 pKa = 5.09DD508 pKa = 5.15DD509 pKa = 4.55NNFYY513 pKa = 10.56IQLGSNNLNSLGNLSLSNPSVTSIIEE539 pKa = 4.48AIEE542 pKa = 4.17LGDD545 pKa = 3.94YY546 pKa = 11.18NGDD549 pKa = 3.68GYY551 pKa = 11.41DD552 pKa = 5.27DD553 pKa = 3.9ILLNTSSNSRR563 pKa = 11.84IYY565 pKa = 10.75LGNEE569 pKa = 3.58EE570 pKa = 4.8GNVNNFLTFNPVGGNSFSDD589 pKa = 3.68YY590 pKa = 10.38TFGSIGDD597 pKa = 3.76IDD599 pKa = 4.27GDD601 pKa = 4.38GYY603 pKa = 11.33QDD605 pKa = 3.21IATGLPQSNFTIQDD619 pKa = 3.29AANGQFTIYY628 pKa = 9.92TNRR631 pKa = 11.84GNQSILPPVTPSLSNFPSNDD651 pKa = 3.5YY652 pKa = 11.1NPTIASYY659 pKa = 10.52GVPSQQSPVPPNFAVYY675 pKa = 9.87QGYY678 pKa = 10.43LYY680 pKa = 7.91MTFKK684 pKa = 11.35GNGNDD689 pKa = 4.67DD690 pKa = 3.86INVMRR695 pKa = 11.84SRR697 pKa = 11.84DD698 pKa = 3.51GNTWEE703 pKa = 4.12NQVVLTSLQTPYY715 pKa = 10.92SPSLVVFEE723 pKa = 5.04DD724 pKa = 3.28KK725 pKa = 10.94LYY727 pKa = 10.81LIHH730 pKa = 6.63TFEE733 pKa = 5.87DD734 pKa = 3.36STIYY738 pKa = 9.1ITPFTGDD745 pKa = 3.76DD746 pKa = 3.35NSISFGEE753 pKa = 4.3YY754 pKa = 8.03TALIADD760 pKa = 5.04AGTSPTPIVFNNKK773 pKa = 9.63LYY775 pKa = 10.47IFYY778 pKa = 10.36IADD781 pKa = 3.62GDD783 pKa = 3.98DD784 pKa = 3.46QINYY788 pKa = 7.19ITLSDD793 pKa = 3.91PANVTEE799 pKa = 4.84FGYY802 pKa = 11.08DD803 pKa = 2.99EE804 pKa = 4.35FNYY807 pKa = 10.12INGQQSFDD815 pKa = 3.37RR816 pKa = 11.84VGATLTPDD824 pKa = 3.49GEE826 pKa = 4.57KK827 pKa = 10.73LVLAYY832 pKa = 9.88KK833 pKa = 10.46NSNKK837 pKa = 9.99ADD839 pKa = 3.23IYY841 pKa = 9.79ITNFDD846 pKa = 3.52GSNWSEE852 pKa = 3.76GKK854 pKa = 9.71IVQDD858 pKa = 3.46SQNNNNTIEE867 pKa = 4.29TTDD870 pKa = 3.74GVSLTTVGNNIYY882 pKa = 10.26LAYY885 pKa = 10.11EE886 pKa = 4.13GTKK889 pKa = 10.69PNDD892 pKa = 2.95LRR894 pKa = 11.84YY895 pKa = 9.69IYY897 pKa = 10.72SQDD900 pKa = 4.24SGNSWEE906 pKa = 4.44NDD908 pKa = 2.82RR909 pKa = 11.84NTPNQLVNGGASLTFFQSSLYY930 pKa = 10.22FGYY933 pKa = 10.44SSQVTSNAPLYY944 pKa = 10.41VSYY947 pKa = 10.96SEE949 pKa = 5.27PIYY952 pKa = 10.02NTNQTQQLGEE962 pKa = 3.92QLYY965 pKa = 11.05SIGDD969 pKa = 3.65FNGDD973 pKa = 3.67GIEE976 pKa = 4.53DD977 pKa = 3.89FAIVAQGYY985 pKa = 9.29IADD988 pKa = 4.75LGIIDD993 pKa = 5.37LSAGLQIRR1001 pKa = 11.84NNQGAILIYY1010 pKa = 10.14YY1011 pKa = 9.31GNANGISNSAKK1022 pKa = 9.64PDD1024 pKa = 3.5VVLVNPPVTDD1034 pKa = 3.73NQSLLYY1040 pKa = 9.97EE1041 pKa = 4.41INRR1044 pKa = 11.84FTAIGDD1050 pKa = 3.82INGDD1054 pKa = 3.47GYY1056 pKa = 11.49DD1057 pKa = 3.94DD1058 pKa = 4.16VAVSSPTTKK1067 pKa = 10.6SNEE1070 pKa = 3.35GSIFVVFGGSSWQSTYY1086 pKa = 11.3SATNPFDD1093 pKa = 5.04LSSLSNNQSNSTTDD1107 pKa = 3.16NSNTNGFLITGLPASQAGISLSGGEE1132 pKa = 4.65DD1133 pKa = 3.46VNGDD1137 pKa = 3.49GFDD1140 pKa = 4.23DD1141 pKa = 4.16FTVGAPGNDD1150 pKa = 3.62DD1151 pKa = 3.83DD1152 pKa = 4.23LTYY1155 pKa = 11.17VIFGSDD1161 pKa = 2.95FTQQVNQTGTIGRR1174 pKa = 11.84DD1175 pKa = 3.15RR1176 pKa = 11.84MIGTPTGEE1184 pKa = 3.63IFIAGEE1190 pKa = 4.16GNDD1193 pKa = 3.74RR1194 pKa = 11.84IYY1196 pKa = 11.46SNGGVDD1202 pKa = 3.4VVYY1205 pKa = 10.93ASVGNDD1211 pKa = 2.95FVTVNDD1217 pKa = 3.87TYY1219 pKa = 11.02FRR1221 pKa = 11.84RR1222 pKa = 11.84LDD1224 pKa = 3.66GGTGIDD1230 pKa = 3.33TLKK1233 pKa = 9.53FTGYY1237 pKa = 10.5NGQNWDD1243 pKa = 4.22LTTLSPGNRR1252 pKa = 11.84LRR1254 pKa = 11.84NFEE1257 pKa = 4.02ILDD1260 pKa = 3.43IKK1262 pKa = 11.13DD1263 pKa = 3.49YY1264 pKa = 11.74GEE1266 pKa = 4.01NTLTLNSLTVTQLSSNNTITVLMDD1290 pKa = 4.47KK1291 pKa = 10.71EE1292 pKa = 4.2DD1293 pKa = 4.0TLNLSADD1300 pKa = 3.86FVNEE1304 pKa = 4.45GIINRR1309 pKa = 11.84NGEE1312 pKa = 4.21TYY1314 pKa = 9.69QKK1316 pKa = 10.91YY1317 pKa = 9.98RR1318 pKa = 11.84IDD1320 pKa = 3.67GATVLILEE1328 pKa = 5.73DD1329 pKa = 3.39IVQTKK1334 pKa = 10.76AFTPLSLSLYY1344 pKa = 8.35NTGVDD1349 pKa = 4.88DD1350 pKa = 6.19SGTPLNNNSTGDD1362 pKa = 3.41PHH1364 pKa = 8.21YY1365 pKa = 10.11EE1366 pKa = 3.92LVSAPDD1372 pKa = 3.95GSSSNILVTNSGDD1385 pKa = 4.16GISGWISPDD1394 pKa = 3.18NNNSDD1399 pKa = 3.6SSPPGDD1405 pKa = 3.39YY1406 pKa = 10.2TYY1408 pKa = 10.19RR1409 pKa = 11.84TTFNLPDD1416 pKa = 3.97TVNLSQFFIDD1426 pKa = 5.46GEE1428 pKa = 4.2WHH1430 pKa = 6.4TDD1432 pKa = 3.41DD1433 pKa = 4.38EE1434 pKa = 5.76GITITLNGIEE1444 pKa = 4.85QYY1446 pKa = 10.73LASNPNFNTDD1456 pKa = 3.91FVPFTINEE1464 pKa = 3.95GFIAGEE1470 pKa = 4.24NILEE1474 pKa = 4.13FTVNNAGTEE1483 pKa = 4.0INPTVLRR1490 pKa = 11.84VKK1492 pKa = 10.47FDD1494 pKa = 3.41NAFDD1498 pKa = 4.39SNIQFSAPSTNIPQAILNPEE1518 pKa = 4.04NVSQVSSANNSVVSIDD1534 pKa = 3.4EE1535 pKa = 4.43SNNITEE1541 pKa = 4.32VSSSLFANTSNNEE1554 pKa = 3.87DD1555 pKa = 3.65STTNLYY1561 pKa = 7.72VTNPTVNEE1569 pKa = 3.67ADD1571 pKa = 3.85GEE1573 pKa = 4.84VKK1575 pKa = 9.33FTIQRR1580 pKa = 11.84TGNLDD1585 pKa = 3.38KK1586 pKa = 10.89YY1587 pKa = 9.41VQIHH1591 pKa = 5.85YY1592 pKa = 7.6VTQDD1596 pKa = 2.69GRR1598 pKa = 11.84AKK1600 pKa = 10.57AGNDD1604 pKa = 3.16YY1605 pKa = 10.93HH1606 pKa = 6.17PTVGKK1611 pKa = 10.51AIFTPGEE1618 pKa = 3.75SSINVSVPLILDD1630 pKa = 3.55DD1631 pKa = 4.75VYY1633 pKa = 11.37TGTKK1637 pKa = 10.25DD1638 pKa = 2.64IGLFVTLEE1646 pKa = 4.36KK1647 pKa = 10.59EE1648 pKa = 4.12SHH1650 pKa = 6.04TPLIDD1655 pKa = 3.36EE1656 pKa = 4.92FNIHH1660 pKa = 7.06ADD1662 pKa = 3.65FNNGIINSWHH1672 pKa = 6.8HH1673 pKa = 5.51IQEE1676 pKa = 4.49EE1677 pKa = 4.72STHH1680 pKa = 6.34PLSLINGEE1688 pKa = 4.03LEE1690 pKa = 4.12FRR1692 pKa = 11.84VTATEE1697 pKa = 4.36GEE1699 pKa = 4.49AEE1701 pKa = 4.22VKK1703 pKa = 10.53LYY1705 pKa = 10.83FDD1707 pKa = 4.84GNHH1710 pKa = 5.83EE1711 pKa = 4.29FNRR1714 pKa = 11.84YY1715 pKa = 9.86YY1716 pKa = 10.29IFNTQNQRR1724 pKa = 11.84YY1725 pKa = 7.95EE1726 pKa = 4.02VFDD1729 pKa = 4.35FNGDD1733 pKa = 3.11TGAEE1737 pKa = 4.21LFDD1740 pKa = 4.89EE1741 pKa = 5.92DD1742 pKa = 5.74DD1743 pKa = 3.96DD1744 pKa = 5.16SNYY1747 pKa = 10.8EE1748 pKa = 4.01GVILHH1753 pKa = 6.2LQDD1756 pKa = 4.48GSEE1759 pKa = 4.17HH1760 pKa = 6.9DD1761 pKa = 4.37LDD1763 pKa = 4.04GARR1766 pKa = 11.84NGVIIKK1772 pKa = 10.28RR1773 pKa = 11.84GFFAGGNPPEE1783 pKa = 4.31TVLNKK1788 pKa = 9.79PMYY1791 pKa = 10.18RR1792 pKa = 11.84FRR1794 pKa = 11.84NSDD1797 pKa = 3.61YY1798 pKa = 10.75DD1799 pKa = 3.54TGAYY1803 pKa = 10.07LYY1805 pKa = 10.44AGEE1808 pKa = 4.86IEE1810 pKa = 4.33SEE1812 pKa = 4.64SIRR1815 pKa = 11.84EE1816 pKa = 3.89NYY1818 pKa = 9.48PNFVEE1823 pKa = 4.5EE1824 pKa = 4.73GFAFYY1829 pKa = 11.16VSTTPQDD1836 pKa = 3.42GLITFNRR1843 pKa = 11.84FQNLDD1848 pKa = 3.41YY1849 pKa = 10.76PGSYY1853 pKa = 10.3LYY1855 pKa = 10.87AEE1857 pKa = 4.38EE1858 pKa = 4.72TEE1860 pKa = 4.35SEE1862 pKa = 4.74SIRR1865 pKa = 11.84EE1866 pKa = 3.94NYY1868 pKa = 9.48PNFVEE1873 pKa = 4.42EE1874 pKa = 5.28GIAFYY1879 pKa = 10.18AYY1881 pKa = 9.46PQGSQMADD1889 pKa = 3.29VISRR1893 pKa = 11.84FQSNDD1898 pKa = 2.94LLGSYY1903 pKa = 10.14LYY1905 pKa = 10.25TSEE1908 pKa = 5.1PEE1910 pKa = 3.8TDD1912 pKa = 3.5NVISNYY1918 pKa = 10.84ANFNLEE1924 pKa = 4.17GIAFEE1929 pKa = 4.58AVVAA1933 pKa = 4.42
Molecular weight: 208.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9Z3X2|K9Z3X2_CYAAP Uncharacterized protein OS=Cyanobacterium aponinum (strain PCC 10605) OX=755178 GN=Cyan10605_0948 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 9.12YY4 pKa = 8.82TLNGTRR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 9.42QKK14 pKa = 8.48RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.37NGRR29 pKa = 11.84KK30 pKa = 9.23VIQARR35 pKa = 11.84RR36 pKa = 11.84NKK38 pKa = 9.03GRR40 pKa = 11.84KK41 pKa = 8.78RR42 pKa = 11.84LAVV45 pKa = 3.41
MM1 pKa = 7.74SKK3 pKa = 9.12YY4 pKa = 8.82TLNGTRR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 9.42QKK14 pKa = 8.48RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.37NGRR29 pKa = 11.84KK30 pKa = 9.23VIQARR35 pKa = 11.84RR36 pKa = 11.84NKK38 pKa = 9.03GRR40 pKa = 11.84KK41 pKa = 8.78RR42 pKa = 11.84LAVV45 pKa = 3.41
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1109571 |
29 |
4183 |
324.9 |
36.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.291 ± 0.04 | 1.001 ± 0.016 |
5.078 ± 0.037 | 6.806 ± 0.044 |
4.327 ± 0.032 | 6.24 ± 0.047 |
1.709 ± 0.023 | 8.363 ± 0.037 |
6.221 ± 0.057 | 10.783 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.823 ± 0.022 | 5.892 ± 0.059 |
4.066 ± 0.027 | 4.807 ± 0.032 |
4.086 ± 0.031 | 6.498 ± 0.036 |
5.384 ± 0.046 | 5.898 ± 0.033 |
1.304 ± 0.019 | 3.424 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
