
Paenibacillus sp. UNCCL117
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Paenibacillus; unclassified Paenibacillus
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
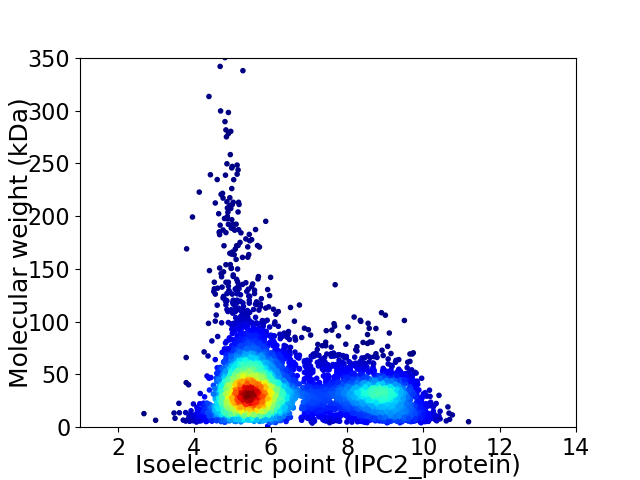
Virtual 2D-PAGE plot for 5982 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1K1LLN7|A0A1K1LLN7_9BACL Putative aminopeptidase FrvX OS=Paenibacillus sp. UNCCL117 OX=1502764 GN=SAMN02799630_00191 PE=3 SV=1
MM1 pKa = 7.38SNKK4 pKa = 9.51NKK6 pKa = 9.96HH7 pKa = 5.52VPPFALKK14 pKa = 10.08AWRR17 pKa = 11.84GLLAASLVIAPASYY31 pKa = 9.22PAAFVHH37 pKa = 7.18ADD39 pKa = 4.08LSPHH43 pKa = 6.23PVHH46 pKa = 7.57DD47 pKa = 3.94IVALPGEE54 pKa = 4.31HH55 pKa = 6.59MKK57 pKa = 10.88LQLKK61 pKa = 10.36DD62 pKa = 4.23LFDD65 pKa = 4.74GNSGVFQLSGTEE77 pKa = 4.03DD78 pKa = 3.68DD79 pKa = 4.18EE80 pKa = 4.96QVVSVNLDD88 pKa = 3.3THH90 pKa = 6.81DD91 pKa = 3.78LQLFFRR97 pKa = 11.84QPGQATFTVTLNGDD111 pKa = 3.49SRR113 pKa = 11.84QVAVNVLEE121 pKa = 4.51AANEE125 pKa = 4.2GPMDD129 pKa = 3.76IGDD132 pKa = 4.14LVRR135 pKa = 11.84FINANPEE142 pKa = 3.92KK143 pKa = 9.82FTDD146 pKa = 3.59RR147 pKa = 11.84SSVSTYY153 pKa = 10.43LQTVAPGEE161 pKa = 4.28VQMNHH166 pKa = 6.26APTVSQATYY175 pKa = 9.91SLRR178 pKa = 11.84YY179 pKa = 7.31TSGYY183 pKa = 7.85TLDD186 pKa = 3.45MSAFFNDD193 pKa = 3.42SDD195 pKa = 4.96GDD197 pKa = 3.84TLDD200 pKa = 3.98YY201 pKa = 11.22VVLPASMNGIEE212 pKa = 4.35ASLSGSVLTLTGTQTAKK229 pKa = 10.65SAFTVYY235 pKa = 11.2AMDD238 pKa = 4.3PKK240 pKa = 11.09GAFASIKK247 pKa = 9.92LLPNTPPAPLSVTGSVYY264 pKa = 10.64AAVYY268 pKa = 7.16EE269 pKa = 4.15QPQAISLSPYY279 pKa = 9.64FIDD282 pKa = 4.05MEE284 pKa = 4.59GDD286 pKa = 3.26EE287 pKa = 4.41LSYY290 pKa = 10.73RR291 pKa = 11.84VAGVTVTDD299 pKa = 3.62YY300 pKa = 10.84TYY302 pKa = 11.25NGQLTPLVPFINPGEE317 pKa = 4.39SILRR321 pKa = 11.84FSGQLAARR329 pKa = 11.84TDD331 pKa = 3.28ISIIAGDD338 pKa = 3.81GQYY341 pKa = 10.78EE342 pKa = 4.31SEE344 pKa = 4.38PFRR347 pKa = 11.84LTLDD351 pKa = 4.31PSPSAYY357 pKa = 9.97NRR359 pKa = 11.84PPVGTPLSLSAYY371 pKa = 9.13KK372 pKa = 10.57NSLLSSVTLSTYY384 pKa = 10.52FHH386 pKa = 7.5DD387 pKa = 5.26PDD389 pKa = 5.2GDD391 pKa = 3.95PLTYY395 pKa = 10.23SVSPSTSGNVTASIANGILSFSGKK419 pKa = 9.43IKK421 pKa = 10.89ANATFTVVATDD432 pKa = 3.03SHH434 pKa = 6.3GASTSAAFTYY444 pKa = 10.34TLDD447 pKa = 3.29NRR449 pKa = 11.84APTGSPVTAVVYY461 pKa = 10.86KK462 pKa = 9.51NTEE465 pKa = 3.75LSSIALASHH474 pKa = 6.45FTDD477 pKa = 5.34PDD479 pKa = 4.53GDD481 pKa = 3.57WLTYY485 pKa = 8.99TVSPPTSGGVTASITSGVLSFSGQLAANASFIVTAKK521 pKa = 10.3DD522 pKa = 3.16AQGAQGSAAFTYY534 pKa = 10.5VLEE537 pKa = 4.23NRR539 pKa = 11.84PPAGTPFTVQVPLYY553 pKa = 6.25TTPDD557 pKa = 3.47SVTLATYY564 pKa = 10.43FSDD567 pKa = 3.97PDD569 pKa = 3.82GDD571 pKa = 3.93ALSYY575 pKa = 10.35TVTPGAPEE583 pKa = 4.16SQNGLAASIIDD594 pKa = 4.74GILSFSGAITGPSEE608 pKa = 4.18FVVRR612 pKa = 11.84AYY614 pKa = 10.68DD615 pKa = 3.48GGGLFGEE622 pKa = 4.79AVFAYY627 pKa = 10.09DD628 pKa = 3.86IANSAPIAAEE638 pKa = 3.69PVVSLNVHH646 pKa = 6.0NEE648 pKa = 3.67VPPIDD653 pKa = 4.24LSANFSDD660 pKa = 4.61PDD662 pKa = 3.78LDD664 pKa = 3.8EE665 pKa = 4.5LTFTVDD671 pKa = 3.11PLEE674 pKa = 4.27AAKK677 pKa = 10.69LPASIEE683 pKa = 3.91NGYY686 pKa = 10.09LLIDD690 pKa = 4.47GMPEE694 pKa = 4.28DD695 pKa = 3.76IVSFTVTAMDD705 pKa = 4.27PSGASASAVYY715 pKa = 10.36QLTPSNTPPIAEE727 pKa = 4.12PHH729 pKa = 5.37TVTGARR735 pKa = 11.84YY736 pKa = 8.06HH737 pKa = 6.71APDD740 pKa = 5.82PIDD743 pKa = 3.4LTEE746 pKa = 4.94FFQDD750 pKa = 4.22DD751 pKa = 4.57DD752 pKa = 4.58GDD754 pKa = 3.93KK755 pKa = 10.59LTFSLTSTEE764 pKa = 4.01EE765 pKa = 4.05SGGLNAYY772 pKa = 9.64IEE774 pKa = 4.65GNHH777 pKa = 6.42LNFSGAQTATAVFTVTGTDD796 pKa = 3.27QMGGTISAMFTIEE809 pKa = 5.77LINQAPTGSSRR820 pKa = 11.84NEE822 pKa = 3.69TVFKK826 pKa = 10.86NDD828 pKa = 3.22EE829 pKa = 4.26LVSLSLDD836 pKa = 4.42DD837 pKa = 5.26YY838 pKa = 11.04FTDD841 pKa = 4.15PDD843 pKa = 4.42GDD845 pKa = 3.79EE846 pKa = 4.07LTYY849 pKa = 10.42TVSPDD854 pKa = 3.06EE855 pKa = 4.75AGGITASIEE864 pKa = 4.19SGVLYY869 pKa = 10.63LSGLIQSDD877 pKa = 3.36ASFTVTATDD886 pKa = 2.77IGGAEE891 pKa = 3.86ATATFSYY898 pKa = 10.98LLANRR903 pKa = 11.84KK904 pKa = 9.27PEE906 pKa = 4.16VVQVPDD912 pKa = 4.93AISTPRR918 pKa = 11.84NGALTSIDD926 pKa = 3.25LSAYY930 pKa = 10.42FSDD933 pKa = 3.96PDD935 pKa = 3.65GDD937 pKa = 3.63EE938 pKa = 5.25LYY940 pKa = 11.14FEE942 pKa = 4.47VLPNEE947 pKa = 4.23EE948 pKa = 4.38VNGITAQINGGVMSFIGSITEE969 pKa = 4.11TTTFTVRR976 pKa = 11.84AYY978 pKa = 10.25DD979 pKa = 3.21HH980 pKa = 6.8SEE982 pKa = 3.88AYY984 pKa = 9.95EE985 pKa = 3.84EE986 pKa = 5.19LEE988 pKa = 4.07FTFLIVNQAPIAGEE1002 pKa = 3.96TLITLQAHH1010 pKa = 5.68NEE1012 pKa = 3.84VGAIHH1017 pKa = 6.91LPSNFSDD1024 pKa = 3.75PDD1026 pKa = 3.6GDD1028 pKa = 3.95KK1029 pKa = 11.33LIFTVDD1035 pKa = 3.41PAVTGDD1041 pKa = 3.89LTTSIQSGNLYY1052 pKa = 8.32ITGVPKK1058 pKa = 10.25EE1059 pKa = 4.05QASFTVTATDD1069 pKa = 3.52EE1070 pKa = 4.42RR1071 pKa = 11.84GEE1073 pKa = 4.47SAWAVYY1079 pKa = 9.01TIVPVNQAPTGSPVTIAGTWNEE1101 pKa = 4.11TPATVDD1107 pKa = 3.52LNDD1110 pKa = 3.56YY1111 pKa = 10.26FQDD1114 pKa = 3.54SDD1116 pKa = 3.65EE1117 pKa = 5.17DD1118 pKa = 3.74EE1119 pKa = 4.78LLFSIAEE1126 pKa = 4.17PTTSGGLTAYY1136 pKa = 10.9LEE1138 pKa = 4.69GGHH1141 pKa = 5.9MLSFTGNQTAEE1152 pKa = 3.74ARR1154 pKa = 11.84FEE1156 pKa = 3.8ITAADD1161 pKa = 3.14IMGGYY1166 pKa = 7.48EE1167 pKa = 4.15TITYY1171 pKa = 9.04TIEE1174 pKa = 3.93LHH1176 pKa = 5.99NLPPVGEE1183 pKa = 4.12VLYY1186 pKa = 10.98LDD1188 pKa = 3.88VGRR1191 pKa = 11.84NSYY1194 pKa = 10.06AAPISLAEE1202 pKa = 4.28LFVDD1206 pKa = 4.99PEE1208 pKa = 4.11NDD1210 pKa = 3.32PLTFEE1215 pKa = 4.73VEE1217 pKa = 4.4TLSSGNVHH1225 pKa = 7.08ASISGNLLMFEE1236 pKa = 4.43GQIEE1240 pKa = 4.12ADD1242 pKa = 2.85ASFRR1246 pKa = 11.84VTATEE1251 pKa = 3.65YY1252 pKa = 10.56RR1253 pKa = 11.84YY1254 pKa = 10.66DD1255 pKa = 3.44QAAPLSATADD1265 pKa = 2.99IHH1267 pKa = 6.85LRR1269 pKa = 11.84AVNSAPEE1276 pKa = 4.04TASPVITDD1284 pKa = 3.87YY1285 pKa = 11.55YY1286 pKa = 10.35HH1287 pKa = 6.32YY1288 pKa = 10.39LYY1290 pKa = 10.86GEE1292 pKa = 4.3AYY1294 pKa = 9.5IYY1296 pKa = 10.1PAVTLAHH1303 pKa = 7.13LFLDD1307 pKa = 4.25PNGDD1311 pKa = 3.82PLTFSLKK1318 pKa = 10.72DD1319 pKa = 3.46GANSGLSEE1327 pKa = 4.12VTVNGVKK1334 pKa = 9.36ATIMPDD1340 pKa = 2.94NEE1342 pKa = 4.66KK1343 pKa = 10.66IMLYY1347 pKa = 10.58FGGNPTNVDD1356 pKa = 3.27EE1357 pKa = 5.7AIEE1360 pKa = 4.26FTVVAADD1367 pKa = 3.75GQGAEE1372 pKa = 4.25TPLTYY1377 pKa = 10.68RR1378 pKa = 11.84LAFNHH1383 pKa = 6.4APEE1386 pKa = 4.65LVDD1389 pKa = 3.98PEE1391 pKa = 4.67GGSEE1395 pKa = 3.96GLAYY1399 pKa = 10.56VYY1401 pKa = 10.04IDD1403 pKa = 3.14HH1404 pKa = 7.24TGTIEE1409 pKa = 5.27DD1410 pKa = 4.67LDD1412 pKa = 4.99LNTLVDD1418 pKa = 5.31DD1419 pKa = 4.98PDD1421 pKa = 4.74GDD1423 pKa = 4.15PLAFTYY1429 pKa = 11.12YY1430 pKa = 10.59EE1431 pKa = 4.68DD1432 pKa = 5.23DD1433 pKa = 3.9SYY1435 pKa = 11.97TDD1437 pKa = 3.71GLSVTLSGSTLSFEE1451 pKa = 4.12GRR1453 pKa = 11.84LNRR1456 pKa = 11.84EE1457 pKa = 3.78TPEE1460 pKa = 4.4PEE1462 pKa = 4.39LALHH1466 pKa = 6.33LQADD1470 pKa = 3.9DD1471 pKa = 4.25GKK1473 pKa = 10.52NGRR1476 pKa = 11.84IDD1478 pKa = 3.72FVLLARR1484 pKa = 11.84YY1485 pKa = 9.39RR1486 pKa = 11.84SIPVASEE1493 pKa = 3.79PEE1495 pKa = 4.05VRR1497 pKa = 11.84VSYY1500 pKa = 10.73LKK1502 pKa = 9.95EE1503 pKa = 3.84QPAGALALSSVYY1515 pKa = 10.75NRR1517 pKa = 11.84FYY1519 pKa = 10.9ATDD1522 pKa = 3.82GNEE1525 pKa = 4.02LFYY1528 pKa = 10.65EE1529 pKa = 4.33IYY1531 pKa = 10.31EE1532 pKa = 4.31PEE1534 pKa = 4.73GEE1536 pKa = 4.28PLPDD1540 pKa = 4.48GYY1542 pKa = 10.53HH1543 pKa = 7.08AEE1545 pKa = 3.93LAYY1548 pKa = 9.91PDD1550 pKa = 4.32GLLHH1554 pKa = 6.05VQLSKK1559 pKa = 11.15DD1560 pKa = 3.2SSVEE1564 pKa = 3.74VEE1566 pKa = 4.27TPYY1569 pKa = 10.75VPVRR1573 pKa = 11.84VRR1575 pKa = 11.84AHH1577 pKa = 6.28GAHH1580 pKa = 6.7GFAEE1584 pKa = 4.11IVYY1587 pKa = 8.81VFQLNQAPASLLPSNVDD1604 pKa = 3.28IYY1606 pKa = 11.11PEE1608 pKa = 3.88IGILNQRR1615 pKa = 11.84EE1616 pKa = 4.21FFMDD1620 pKa = 3.98PDD1622 pKa = 4.91GGDD1625 pKa = 3.2SFTVEE1630 pKa = 3.63MSITDD1635 pKa = 3.3RR1636 pKa = 11.84GYY1638 pKa = 10.86DD1639 pKa = 3.57GQNYY1643 pKa = 7.99WLGITGGSYY1652 pKa = 10.63EE1653 pKa = 5.02LIGLNPYY1660 pKa = 10.56LEE1662 pKa = 5.53DD1663 pKa = 3.94GLNGIISFTATDD1675 pKa = 3.15NHH1677 pKa = 5.83TQSSQQTVEE1686 pKa = 3.8FHH1688 pKa = 5.7KK1689 pKa = 9.53TAASEE1694 pKa = 4.3VIQMPDD1700 pKa = 2.73KK1701 pKa = 10.54RR1702 pKa = 11.84LSAGTEE1708 pKa = 3.91LTLKK1712 pKa = 10.54DD1713 pKa = 3.47LSTRR1717 pKa = 11.84YY1718 pKa = 10.41SLDD1721 pKa = 3.55TIHH1724 pKa = 7.1KK1725 pKa = 8.97FKK1727 pKa = 10.97ASSDD1731 pKa = 3.84DD1732 pKa = 3.7PEE1734 pKa = 6.17GMLVSISNDD1743 pKa = 3.45GQDD1746 pKa = 3.13LTLAAEE1752 pKa = 4.09HH1753 pKa = 6.75AGTYY1757 pKa = 10.0RR1758 pKa = 11.84ITLIGTAADD1767 pKa = 3.46DD1768 pKa = 3.85TGFIDD1773 pKa = 3.5QFYY1776 pKa = 8.91VTVTEE1781 pKa = 4.17PEE1783 pKa = 3.99IFGSSLTISNPFGTEE1798 pKa = 3.8AIVNRR1803 pKa = 11.84DD1804 pKa = 3.3SVEE1807 pKa = 4.01SEE1809 pKa = 3.95NANFAAVLEE1818 pKa = 4.86DD1819 pKa = 3.91EE1820 pKa = 5.35ADD1822 pKa = 3.86PDD1824 pKa = 4.13LLQLQVSVDD1833 pKa = 3.17EE1834 pKa = 4.78GAAFLEE1840 pKa = 3.97NTYY1843 pKa = 10.71FILYY1847 pKa = 10.33SNGAIYY1853 pKa = 10.16KK1854 pKa = 9.87IRR1856 pKa = 11.84FYY1858 pKa = 11.56SNTTAA1863 pKa = 4.53
MM1 pKa = 7.38SNKK4 pKa = 9.51NKK6 pKa = 9.96HH7 pKa = 5.52VPPFALKK14 pKa = 10.08AWRR17 pKa = 11.84GLLAASLVIAPASYY31 pKa = 9.22PAAFVHH37 pKa = 7.18ADD39 pKa = 4.08LSPHH43 pKa = 6.23PVHH46 pKa = 7.57DD47 pKa = 3.94IVALPGEE54 pKa = 4.31HH55 pKa = 6.59MKK57 pKa = 10.88LQLKK61 pKa = 10.36DD62 pKa = 4.23LFDD65 pKa = 4.74GNSGVFQLSGTEE77 pKa = 4.03DD78 pKa = 3.68DD79 pKa = 4.18EE80 pKa = 4.96QVVSVNLDD88 pKa = 3.3THH90 pKa = 6.81DD91 pKa = 3.78LQLFFRR97 pKa = 11.84QPGQATFTVTLNGDD111 pKa = 3.49SRR113 pKa = 11.84QVAVNVLEE121 pKa = 4.51AANEE125 pKa = 4.2GPMDD129 pKa = 3.76IGDD132 pKa = 4.14LVRR135 pKa = 11.84FINANPEE142 pKa = 3.92KK143 pKa = 9.82FTDD146 pKa = 3.59RR147 pKa = 11.84SSVSTYY153 pKa = 10.43LQTVAPGEE161 pKa = 4.28VQMNHH166 pKa = 6.26APTVSQATYY175 pKa = 9.91SLRR178 pKa = 11.84YY179 pKa = 7.31TSGYY183 pKa = 7.85TLDD186 pKa = 3.45MSAFFNDD193 pKa = 3.42SDD195 pKa = 4.96GDD197 pKa = 3.84TLDD200 pKa = 3.98YY201 pKa = 11.22VVLPASMNGIEE212 pKa = 4.35ASLSGSVLTLTGTQTAKK229 pKa = 10.65SAFTVYY235 pKa = 11.2AMDD238 pKa = 4.3PKK240 pKa = 11.09GAFASIKK247 pKa = 9.92LLPNTPPAPLSVTGSVYY264 pKa = 10.64AAVYY268 pKa = 7.16EE269 pKa = 4.15QPQAISLSPYY279 pKa = 9.64FIDD282 pKa = 4.05MEE284 pKa = 4.59GDD286 pKa = 3.26EE287 pKa = 4.41LSYY290 pKa = 10.73RR291 pKa = 11.84VAGVTVTDD299 pKa = 3.62YY300 pKa = 10.84TYY302 pKa = 11.25NGQLTPLVPFINPGEE317 pKa = 4.39SILRR321 pKa = 11.84FSGQLAARR329 pKa = 11.84TDD331 pKa = 3.28ISIIAGDD338 pKa = 3.81GQYY341 pKa = 10.78EE342 pKa = 4.31SEE344 pKa = 4.38PFRR347 pKa = 11.84LTLDD351 pKa = 4.31PSPSAYY357 pKa = 9.97NRR359 pKa = 11.84PPVGTPLSLSAYY371 pKa = 9.13KK372 pKa = 10.57NSLLSSVTLSTYY384 pKa = 10.52FHH386 pKa = 7.5DD387 pKa = 5.26PDD389 pKa = 5.2GDD391 pKa = 3.95PLTYY395 pKa = 10.23SVSPSTSGNVTASIANGILSFSGKK419 pKa = 9.43IKK421 pKa = 10.89ANATFTVVATDD432 pKa = 3.03SHH434 pKa = 6.3GASTSAAFTYY444 pKa = 10.34TLDD447 pKa = 3.29NRR449 pKa = 11.84APTGSPVTAVVYY461 pKa = 10.86KK462 pKa = 9.51NTEE465 pKa = 3.75LSSIALASHH474 pKa = 6.45FTDD477 pKa = 5.34PDD479 pKa = 4.53GDD481 pKa = 3.57WLTYY485 pKa = 8.99TVSPPTSGGVTASITSGVLSFSGQLAANASFIVTAKK521 pKa = 10.3DD522 pKa = 3.16AQGAQGSAAFTYY534 pKa = 10.5VLEE537 pKa = 4.23NRR539 pKa = 11.84PPAGTPFTVQVPLYY553 pKa = 6.25TTPDD557 pKa = 3.47SVTLATYY564 pKa = 10.43FSDD567 pKa = 3.97PDD569 pKa = 3.82GDD571 pKa = 3.93ALSYY575 pKa = 10.35TVTPGAPEE583 pKa = 4.16SQNGLAASIIDD594 pKa = 4.74GILSFSGAITGPSEE608 pKa = 4.18FVVRR612 pKa = 11.84AYY614 pKa = 10.68DD615 pKa = 3.48GGGLFGEE622 pKa = 4.79AVFAYY627 pKa = 10.09DD628 pKa = 3.86IANSAPIAAEE638 pKa = 3.69PVVSLNVHH646 pKa = 6.0NEE648 pKa = 3.67VPPIDD653 pKa = 4.24LSANFSDD660 pKa = 4.61PDD662 pKa = 3.78LDD664 pKa = 3.8EE665 pKa = 4.5LTFTVDD671 pKa = 3.11PLEE674 pKa = 4.27AAKK677 pKa = 10.69LPASIEE683 pKa = 3.91NGYY686 pKa = 10.09LLIDD690 pKa = 4.47GMPEE694 pKa = 4.28DD695 pKa = 3.76IVSFTVTAMDD705 pKa = 4.27PSGASASAVYY715 pKa = 10.36QLTPSNTPPIAEE727 pKa = 4.12PHH729 pKa = 5.37TVTGARR735 pKa = 11.84YY736 pKa = 8.06HH737 pKa = 6.71APDD740 pKa = 5.82PIDD743 pKa = 3.4LTEE746 pKa = 4.94FFQDD750 pKa = 4.22DD751 pKa = 4.57DD752 pKa = 4.58GDD754 pKa = 3.93KK755 pKa = 10.59LTFSLTSTEE764 pKa = 4.01EE765 pKa = 4.05SGGLNAYY772 pKa = 9.64IEE774 pKa = 4.65GNHH777 pKa = 6.42LNFSGAQTATAVFTVTGTDD796 pKa = 3.27QMGGTISAMFTIEE809 pKa = 5.77LINQAPTGSSRR820 pKa = 11.84NEE822 pKa = 3.69TVFKK826 pKa = 10.86NDD828 pKa = 3.22EE829 pKa = 4.26LVSLSLDD836 pKa = 4.42DD837 pKa = 5.26YY838 pKa = 11.04FTDD841 pKa = 4.15PDD843 pKa = 4.42GDD845 pKa = 3.79EE846 pKa = 4.07LTYY849 pKa = 10.42TVSPDD854 pKa = 3.06EE855 pKa = 4.75AGGITASIEE864 pKa = 4.19SGVLYY869 pKa = 10.63LSGLIQSDD877 pKa = 3.36ASFTVTATDD886 pKa = 2.77IGGAEE891 pKa = 3.86ATATFSYY898 pKa = 10.98LLANRR903 pKa = 11.84KK904 pKa = 9.27PEE906 pKa = 4.16VVQVPDD912 pKa = 4.93AISTPRR918 pKa = 11.84NGALTSIDD926 pKa = 3.25LSAYY930 pKa = 10.42FSDD933 pKa = 3.96PDD935 pKa = 3.65GDD937 pKa = 3.63EE938 pKa = 5.25LYY940 pKa = 11.14FEE942 pKa = 4.47VLPNEE947 pKa = 4.23EE948 pKa = 4.38VNGITAQINGGVMSFIGSITEE969 pKa = 4.11TTTFTVRR976 pKa = 11.84AYY978 pKa = 10.25DD979 pKa = 3.21HH980 pKa = 6.8SEE982 pKa = 3.88AYY984 pKa = 9.95EE985 pKa = 3.84EE986 pKa = 5.19LEE988 pKa = 4.07FTFLIVNQAPIAGEE1002 pKa = 3.96TLITLQAHH1010 pKa = 5.68NEE1012 pKa = 3.84VGAIHH1017 pKa = 6.91LPSNFSDD1024 pKa = 3.75PDD1026 pKa = 3.6GDD1028 pKa = 3.95KK1029 pKa = 11.33LIFTVDD1035 pKa = 3.41PAVTGDD1041 pKa = 3.89LTTSIQSGNLYY1052 pKa = 8.32ITGVPKK1058 pKa = 10.25EE1059 pKa = 4.05QASFTVTATDD1069 pKa = 3.52EE1070 pKa = 4.42RR1071 pKa = 11.84GEE1073 pKa = 4.47SAWAVYY1079 pKa = 9.01TIVPVNQAPTGSPVTIAGTWNEE1101 pKa = 4.11TPATVDD1107 pKa = 3.52LNDD1110 pKa = 3.56YY1111 pKa = 10.26FQDD1114 pKa = 3.54SDD1116 pKa = 3.65EE1117 pKa = 5.17DD1118 pKa = 3.74EE1119 pKa = 4.78LLFSIAEE1126 pKa = 4.17PTTSGGLTAYY1136 pKa = 10.9LEE1138 pKa = 4.69GGHH1141 pKa = 5.9MLSFTGNQTAEE1152 pKa = 3.74ARR1154 pKa = 11.84FEE1156 pKa = 3.8ITAADD1161 pKa = 3.14IMGGYY1166 pKa = 7.48EE1167 pKa = 4.15TITYY1171 pKa = 9.04TIEE1174 pKa = 3.93LHH1176 pKa = 5.99NLPPVGEE1183 pKa = 4.12VLYY1186 pKa = 10.98LDD1188 pKa = 3.88VGRR1191 pKa = 11.84NSYY1194 pKa = 10.06AAPISLAEE1202 pKa = 4.28LFVDD1206 pKa = 4.99PEE1208 pKa = 4.11NDD1210 pKa = 3.32PLTFEE1215 pKa = 4.73VEE1217 pKa = 4.4TLSSGNVHH1225 pKa = 7.08ASISGNLLMFEE1236 pKa = 4.43GQIEE1240 pKa = 4.12ADD1242 pKa = 2.85ASFRR1246 pKa = 11.84VTATEE1251 pKa = 3.65YY1252 pKa = 10.56RR1253 pKa = 11.84YY1254 pKa = 10.66DD1255 pKa = 3.44QAAPLSATADD1265 pKa = 2.99IHH1267 pKa = 6.85LRR1269 pKa = 11.84AVNSAPEE1276 pKa = 4.04TASPVITDD1284 pKa = 3.87YY1285 pKa = 11.55YY1286 pKa = 10.35HH1287 pKa = 6.32YY1288 pKa = 10.39LYY1290 pKa = 10.86GEE1292 pKa = 4.3AYY1294 pKa = 9.5IYY1296 pKa = 10.1PAVTLAHH1303 pKa = 7.13LFLDD1307 pKa = 4.25PNGDD1311 pKa = 3.82PLTFSLKK1318 pKa = 10.72DD1319 pKa = 3.46GANSGLSEE1327 pKa = 4.12VTVNGVKK1334 pKa = 9.36ATIMPDD1340 pKa = 2.94NEE1342 pKa = 4.66KK1343 pKa = 10.66IMLYY1347 pKa = 10.58FGGNPTNVDD1356 pKa = 3.27EE1357 pKa = 5.7AIEE1360 pKa = 4.26FTVVAADD1367 pKa = 3.75GQGAEE1372 pKa = 4.25TPLTYY1377 pKa = 10.68RR1378 pKa = 11.84LAFNHH1383 pKa = 6.4APEE1386 pKa = 4.65LVDD1389 pKa = 3.98PEE1391 pKa = 4.67GGSEE1395 pKa = 3.96GLAYY1399 pKa = 10.56VYY1401 pKa = 10.04IDD1403 pKa = 3.14HH1404 pKa = 7.24TGTIEE1409 pKa = 5.27DD1410 pKa = 4.67LDD1412 pKa = 4.99LNTLVDD1418 pKa = 5.31DD1419 pKa = 4.98PDD1421 pKa = 4.74GDD1423 pKa = 4.15PLAFTYY1429 pKa = 11.12YY1430 pKa = 10.59EE1431 pKa = 4.68DD1432 pKa = 5.23DD1433 pKa = 3.9SYY1435 pKa = 11.97TDD1437 pKa = 3.71GLSVTLSGSTLSFEE1451 pKa = 4.12GRR1453 pKa = 11.84LNRR1456 pKa = 11.84EE1457 pKa = 3.78TPEE1460 pKa = 4.4PEE1462 pKa = 4.39LALHH1466 pKa = 6.33LQADD1470 pKa = 3.9DD1471 pKa = 4.25GKK1473 pKa = 10.52NGRR1476 pKa = 11.84IDD1478 pKa = 3.72FVLLARR1484 pKa = 11.84YY1485 pKa = 9.39RR1486 pKa = 11.84SIPVASEE1493 pKa = 3.79PEE1495 pKa = 4.05VRR1497 pKa = 11.84VSYY1500 pKa = 10.73LKK1502 pKa = 9.95EE1503 pKa = 3.84QPAGALALSSVYY1515 pKa = 10.75NRR1517 pKa = 11.84FYY1519 pKa = 10.9ATDD1522 pKa = 3.82GNEE1525 pKa = 4.02LFYY1528 pKa = 10.65EE1529 pKa = 4.33IYY1531 pKa = 10.31EE1532 pKa = 4.31PEE1534 pKa = 4.73GEE1536 pKa = 4.28PLPDD1540 pKa = 4.48GYY1542 pKa = 10.53HH1543 pKa = 7.08AEE1545 pKa = 3.93LAYY1548 pKa = 9.91PDD1550 pKa = 4.32GLLHH1554 pKa = 6.05VQLSKK1559 pKa = 11.15DD1560 pKa = 3.2SSVEE1564 pKa = 3.74VEE1566 pKa = 4.27TPYY1569 pKa = 10.75VPVRR1573 pKa = 11.84VRR1575 pKa = 11.84AHH1577 pKa = 6.28GAHH1580 pKa = 6.7GFAEE1584 pKa = 4.11IVYY1587 pKa = 8.81VFQLNQAPASLLPSNVDD1604 pKa = 3.28IYY1606 pKa = 11.11PEE1608 pKa = 3.88IGILNQRR1615 pKa = 11.84EE1616 pKa = 4.21FFMDD1620 pKa = 3.98PDD1622 pKa = 4.91GGDD1625 pKa = 3.2SFTVEE1630 pKa = 3.63MSITDD1635 pKa = 3.3RR1636 pKa = 11.84GYY1638 pKa = 10.86DD1639 pKa = 3.57GQNYY1643 pKa = 7.99WLGITGGSYY1652 pKa = 10.63EE1653 pKa = 5.02LIGLNPYY1660 pKa = 10.56LEE1662 pKa = 5.53DD1663 pKa = 3.94GLNGIISFTATDD1675 pKa = 3.15NHH1677 pKa = 5.83TQSSQQTVEE1686 pKa = 3.8FHH1688 pKa = 5.7KK1689 pKa = 9.53TAASEE1694 pKa = 4.3VIQMPDD1700 pKa = 2.73KK1701 pKa = 10.54RR1702 pKa = 11.84LSAGTEE1708 pKa = 3.91LTLKK1712 pKa = 10.54DD1713 pKa = 3.47LSTRR1717 pKa = 11.84YY1718 pKa = 10.41SLDD1721 pKa = 3.55TIHH1724 pKa = 7.1KK1725 pKa = 8.97FKK1727 pKa = 10.97ASSDD1731 pKa = 3.84DD1732 pKa = 3.7PEE1734 pKa = 6.17GMLVSISNDD1743 pKa = 3.45GQDD1746 pKa = 3.13LTLAAEE1752 pKa = 4.09HH1753 pKa = 6.75AGTYY1757 pKa = 10.0RR1758 pKa = 11.84ITLIGTAADD1767 pKa = 3.46DD1768 pKa = 3.85TGFIDD1773 pKa = 3.5QFYY1776 pKa = 8.91VTVTEE1781 pKa = 4.17PEE1783 pKa = 3.99IFGSSLTISNPFGTEE1798 pKa = 3.8AIVNRR1803 pKa = 11.84DD1804 pKa = 3.3SVEE1807 pKa = 4.01SEE1809 pKa = 3.95NANFAAVLEE1818 pKa = 4.86DD1819 pKa = 3.91EE1820 pKa = 5.35ADD1822 pKa = 3.86PDD1824 pKa = 4.13LLQLQVSVDD1833 pKa = 3.17EE1834 pKa = 4.78GAAFLEE1840 pKa = 3.97NTYY1843 pKa = 10.71FILYY1847 pKa = 10.33SNGAIYY1853 pKa = 10.16KK1854 pKa = 9.87IRR1856 pKa = 11.84FYY1858 pKa = 11.56SNTTAA1863 pKa = 4.53
Molecular weight: 199.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1K1Q3S0|A0A1K1Q3S0_9BACL Ribosomal RNA large subunit methyltransferase H OS=Paenibacillus sp. UNCCL117 OX=1502764 GN=rlmH PE=3 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84PTFKK6 pKa = 10.87PNVRR10 pKa = 11.84KK11 pKa = 9.84RR12 pKa = 11.84KK13 pKa = 8.89KK14 pKa = 8.64VHH16 pKa = 5.57GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 10.18NGRR28 pKa = 11.84KK29 pKa = 8.87VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.79VLSAA44 pKa = 4.05
MM1 pKa = 7.9RR2 pKa = 11.84PTFKK6 pKa = 10.87PNVRR10 pKa = 11.84KK11 pKa = 9.84RR12 pKa = 11.84KK13 pKa = 8.89KK14 pKa = 8.64VHH16 pKa = 5.57GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 10.18NGRR28 pKa = 11.84KK29 pKa = 8.87VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.79VLSAA44 pKa = 4.05
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2115940 |
9 |
3347 |
353.7 |
39.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.233 ± 0.039 | 0.72 ± 0.01 |
5.063 ± 0.024 | 6.653 ± 0.036 |
3.954 ± 0.023 | 7.804 ± 0.034 |
2.045 ± 0.017 | 5.921 ± 0.028 |
4.883 ± 0.032 | 10.262 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.704 ± 0.017 | 3.384 ± 0.026 |
4.347 ± 0.02 | 3.906 ± 0.02 |
5.582 ± 0.033 | 6.221 ± 0.025 |
5.415 ± 0.029 | 7.022 ± 0.026 |
1.354 ± 0.012 | 3.525 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
