
Shahe isopoda virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.75
Get precalculated fractions of proteins
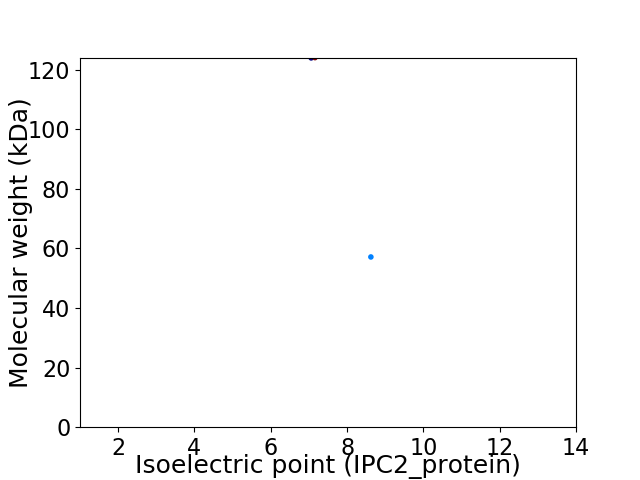
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KL89|A0A1L3KL89_9VIRU Uncharacterized protein OS=Shahe isopoda virus 3 OX=1923423 PE=4 SV=1
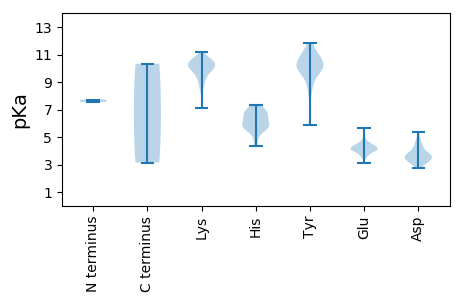
MM1 pKa = 7.55AAPVGANYY9 pKa = 7.49FTQAALNTLAYY20 pKa = 9.94VAGIPGLDD28 pKa = 3.49PRR30 pKa = 11.84DD31 pKa = 4.74LIMQGHH37 pKa = 7.07PEE39 pKa = 3.91LFEE42 pKa = 4.24YY43 pKa = 10.48EE44 pKa = 4.49CLALPGMTCGEE55 pKa = 4.18SALRR59 pKa = 11.84LASAAVWSEE68 pKa = 3.85TNLKK72 pKa = 10.13KK73 pKa = 10.34KK74 pKa = 10.38RR75 pKa = 11.84QDD77 pKa = 3.13RR78 pKa = 11.84DD79 pKa = 3.41PPPFLLHH86 pKa = 6.23GVAGALTTLQMVRR99 pKa = 11.84RR100 pKa = 11.84LNRR103 pKa = 11.84LSMGSFPIYY112 pKa = 9.95RR113 pKa = 11.84YY114 pKa = 10.44AGLDD118 pKa = 3.32VLGAPMFDD126 pKa = 4.79LISGKK131 pKa = 9.25PRR133 pKa = 11.84ADD135 pKa = 3.17NYY137 pKa = 11.26ACVFVPQNLVLVPAFPVAHH156 pKa = 5.82WTFTTILPPPDD167 pKa = 3.33VPDD170 pKa = 3.75RR171 pKa = 11.84LVVGLPTLIYY181 pKa = 10.03TRR183 pKa = 11.84LPLAPQMDD191 pKa = 4.69PVPGQIVFWRR201 pKa = 11.84ARR203 pKa = 11.84VTPEE207 pKa = 3.41NAEE210 pKa = 3.98AFAAAAAVGVACKK223 pKa = 10.51CPWIQVCDD231 pKa = 4.09HH232 pKa = 5.8EE233 pKa = 4.75QAWLAVNPFFSRR245 pKa = 11.84VTCADD250 pKa = 3.64SACMEE255 pKa = 4.27GSSYY259 pKa = 11.87AMVLKK264 pKa = 9.61TLSPDD269 pKa = 4.22FILRR273 pKa = 11.84RR274 pKa = 11.84IPNNGVKK281 pKa = 9.92VLQHH285 pKa = 4.82NTVLKK290 pKa = 9.67EE291 pKa = 4.05EE292 pKa = 4.41NVFGCPTIRR301 pKa = 11.84SEE303 pKa = 5.14RR304 pKa = 11.84FFNTLATAFGSLEE317 pKa = 3.98LQPYY321 pKa = 9.43QSVAEE326 pKa = 4.0RR327 pKa = 11.84DD328 pKa = 3.08IYY330 pKa = 10.29PYY332 pKa = 10.48RR333 pKa = 11.84FRR335 pKa = 11.84VYY337 pKa = 9.24TYY339 pKa = 10.05GRR341 pKa = 11.84CVEE344 pKa = 4.27HH345 pKa = 5.83ATMYY349 pKa = 11.68NMFKK353 pKa = 9.83TLCLDD358 pKa = 3.66TASMLSPYY366 pKa = 9.18TFQRR370 pKa = 11.84EE371 pKa = 4.24FEE373 pKa = 4.37VEE375 pKa = 3.99PVAPAKK381 pKa = 10.26PWGEE385 pKa = 4.21CGDD388 pKa = 3.7LTLRR392 pKa = 11.84LPRR395 pKa = 11.84LNEE398 pKa = 3.66LLQRR402 pKa = 11.84LAVRR406 pKa = 11.84KK407 pKa = 8.99EE408 pKa = 3.63VTADD412 pKa = 3.43VVIDD416 pKa = 3.59TVRR419 pKa = 11.84RR420 pKa = 11.84LAHH423 pKa = 5.31EE424 pKa = 4.54EE425 pKa = 3.73KK426 pKa = 8.48WTTEE430 pKa = 3.1IDD432 pKa = 3.45RR433 pKa = 11.84DD434 pKa = 4.04EE435 pKa = 4.72FEE437 pKa = 4.48LWLSRR442 pKa = 11.84VVTEE446 pKa = 4.3AGEE449 pKa = 4.09RR450 pKa = 11.84AQPLTLAYY458 pKa = 9.78TNGQCWTCGTFGKK471 pKa = 7.08THH473 pKa = 5.6RR474 pKa = 11.84HH475 pKa = 4.33EE476 pKa = 4.95CKK478 pKa = 9.4ACKK481 pKa = 9.55RR482 pKa = 11.84RR483 pKa = 11.84ARR485 pKa = 11.84NRR487 pKa = 11.84VPDD490 pKa = 4.11RR491 pKa = 11.84PLSDD495 pKa = 4.21AFATHH500 pKa = 6.35MGFFPLWSEE509 pKa = 4.3RR510 pKa = 11.84FMLPNMAFKK519 pKa = 10.7RR520 pKa = 11.84GTQVRR525 pKa = 11.84VGKK528 pKa = 10.42AGNRR532 pKa = 11.84VMTTRR537 pKa = 11.84AEE539 pKa = 4.09VNAWLSRR546 pKa = 11.84QRR548 pKa = 11.84VDD550 pKa = 2.94VSQRR554 pKa = 11.84GRR556 pKa = 11.84LCGPMFCNQVPKK568 pKa = 10.43CYY570 pKa = 9.92PKK572 pKa = 10.44GQAVALSAFLVRR584 pKa = 11.84LGGTRR589 pKa = 11.84PYY591 pKa = 10.54PDD593 pKa = 3.6TFPCQMCASDD603 pKa = 3.58GHH605 pKa = 5.7EE606 pKa = 4.2EE607 pKa = 4.23CTKK610 pKa = 10.64QCTPDD615 pKa = 3.47EE616 pKa = 4.45LVFAPNNVRR625 pKa = 11.84PEE627 pKa = 3.79VKK629 pKa = 10.07LAYY632 pKa = 9.95DD633 pKa = 3.66LLYY636 pKa = 10.4TVFSQYY642 pKa = 10.86VLYY645 pKa = 10.92GGLTSASCPLKK656 pKa = 10.53PEE658 pKa = 3.84SRR660 pKa = 11.84TEE662 pKa = 4.01FLSHH666 pKa = 7.04FAGEE670 pKa = 4.46KK671 pKa = 8.52LLKK674 pKa = 9.71MVEE677 pKa = 4.06ALAMINDD684 pKa = 3.71GWLEE688 pKa = 3.97NMDD691 pKa = 4.21DD692 pKa = 3.42EE693 pKa = 4.61AVRR696 pKa = 11.84VRR698 pKa = 11.84FTGFTKK704 pKa = 10.62AEE706 pKa = 3.7KK707 pKa = 10.49SYY709 pKa = 11.01DD710 pKa = 3.74FEE712 pKa = 4.47WTGDD716 pKa = 3.66CLILKK721 pKa = 10.22KK722 pKa = 10.28EE723 pKa = 3.98GKK725 pKa = 9.28PRR727 pKa = 11.84FICSPDD733 pKa = 3.4PVMLYY738 pKa = 10.05YY739 pKa = 10.86LGRR742 pKa = 11.84YY743 pKa = 5.89THH745 pKa = 6.55VQTKK749 pKa = 8.5WLSARR754 pKa = 11.84FLPCSRR760 pKa = 11.84MFYY763 pKa = 10.22AGCATPEE770 pKa = 4.48DD771 pKa = 3.92MNVWLNYY778 pKa = 8.27TAGDD782 pKa = 3.73AQYY785 pKa = 10.46MNTIVDD791 pKa = 4.42DD792 pKa = 4.29VSAMDD797 pKa = 3.75SSFTKK802 pKa = 10.24TLLDD806 pKa = 3.3FHH808 pKa = 6.69EE809 pKa = 5.68RR810 pKa = 11.84IRR812 pKa = 11.84DD813 pKa = 3.48LQFPHH818 pKa = 6.31MAAHH822 pKa = 6.41TKK824 pKa = 9.84AAFRR828 pKa = 11.84GEE830 pKa = 3.58EE831 pKa = 4.14RR832 pKa = 11.84FWVRR836 pKa = 11.84IGEE839 pKa = 3.83IRR841 pKa = 11.84AFVEE845 pKa = 4.1WVNASGVPDD854 pKa = 3.7TSYY857 pKa = 11.76KK858 pKa = 9.03NTAPCLPLRR867 pKa = 11.84VFAIVHH873 pKa = 6.78AIWGMIGQPFEE884 pKa = 4.2LTIEE888 pKa = 4.34RR889 pKa = 11.84YY890 pKa = 9.95YY891 pKa = 11.31LVLDD895 pKa = 4.86GIYY898 pKa = 10.17TSAAGDD904 pKa = 3.79DD905 pKa = 3.85GLTRR909 pKa = 11.84VPTHH913 pKa = 5.89MFGVDD918 pKa = 3.1TSSPQFKK925 pKa = 9.69MGYY928 pKa = 8.31CKK930 pKa = 10.26AWSFFGFNVKK940 pKa = 9.79VDD942 pKa = 3.6IVPPCRR948 pKa = 11.84WRR950 pKa = 11.84MATYY954 pKa = 10.06LAQRR958 pKa = 11.84PVWVGHH964 pKa = 5.75RR965 pKa = 11.84YY966 pKa = 8.37EE967 pKa = 4.4WAPEE971 pKa = 3.46PARR974 pKa = 11.84RR975 pKa = 11.84LRR977 pKa = 11.84GIFWQIDD984 pKa = 3.77CSLHH988 pKa = 5.63PIAWGRR994 pKa = 11.84GIATQLLQQAKK1005 pKa = 8.68ALPVVSHH1012 pKa = 5.55VCQWYY1017 pKa = 9.05LARR1020 pKa = 11.84TSGPAIISAATAASHH1035 pKa = 6.57EE1036 pKa = 4.18YY1037 pKa = 10.78SPFHH1041 pKa = 6.49GAVCSGDD1048 pKa = 3.15INEE1051 pKa = 4.66RR1052 pKa = 11.84GVSEE1056 pKa = 4.21FCVDD1060 pKa = 3.43YY1061 pKa = 10.93NIPRR1065 pKa = 11.84AEE1067 pKa = 4.15LDD1069 pKa = 3.4RR1070 pKa = 11.84FCRR1073 pKa = 11.84LLEE1076 pKa = 4.09LVPSVLVNLNSFVLDD1091 pKa = 4.21RR1092 pKa = 11.84IYY1094 pKa = 11.24AEE1096 pKa = 4.3EE1097 pKa = 4.06SS1098 pKa = 3.15
MM1 pKa = 7.55AAPVGANYY9 pKa = 7.49FTQAALNTLAYY20 pKa = 9.94VAGIPGLDD28 pKa = 3.49PRR30 pKa = 11.84DD31 pKa = 4.74LIMQGHH37 pKa = 7.07PEE39 pKa = 3.91LFEE42 pKa = 4.24YY43 pKa = 10.48EE44 pKa = 4.49CLALPGMTCGEE55 pKa = 4.18SALRR59 pKa = 11.84LASAAVWSEE68 pKa = 3.85TNLKK72 pKa = 10.13KK73 pKa = 10.34KK74 pKa = 10.38RR75 pKa = 11.84QDD77 pKa = 3.13RR78 pKa = 11.84DD79 pKa = 3.41PPPFLLHH86 pKa = 6.23GVAGALTTLQMVRR99 pKa = 11.84RR100 pKa = 11.84LNRR103 pKa = 11.84LSMGSFPIYY112 pKa = 9.95RR113 pKa = 11.84YY114 pKa = 10.44AGLDD118 pKa = 3.32VLGAPMFDD126 pKa = 4.79LISGKK131 pKa = 9.25PRR133 pKa = 11.84ADD135 pKa = 3.17NYY137 pKa = 11.26ACVFVPQNLVLVPAFPVAHH156 pKa = 5.82WTFTTILPPPDD167 pKa = 3.33VPDD170 pKa = 3.75RR171 pKa = 11.84LVVGLPTLIYY181 pKa = 10.03TRR183 pKa = 11.84LPLAPQMDD191 pKa = 4.69PVPGQIVFWRR201 pKa = 11.84ARR203 pKa = 11.84VTPEE207 pKa = 3.41NAEE210 pKa = 3.98AFAAAAAVGVACKK223 pKa = 10.51CPWIQVCDD231 pKa = 4.09HH232 pKa = 5.8EE233 pKa = 4.75QAWLAVNPFFSRR245 pKa = 11.84VTCADD250 pKa = 3.64SACMEE255 pKa = 4.27GSSYY259 pKa = 11.87AMVLKK264 pKa = 9.61TLSPDD269 pKa = 4.22FILRR273 pKa = 11.84RR274 pKa = 11.84IPNNGVKK281 pKa = 9.92VLQHH285 pKa = 4.82NTVLKK290 pKa = 9.67EE291 pKa = 4.05EE292 pKa = 4.41NVFGCPTIRR301 pKa = 11.84SEE303 pKa = 5.14RR304 pKa = 11.84FFNTLATAFGSLEE317 pKa = 3.98LQPYY321 pKa = 9.43QSVAEE326 pKa = 4.0RR327 pKa = 11.84DD328 pKa = 3.08IYY330 pKa = 10.29PYY332 pKa = 10.48RR333 pKa = 11.84FRR335 pKa = 11.84VYY337 pKa = 9.24TYY339 pKa = 10.05GRR341 pKa = 11.84CVEE344 pKa = 4.27HH345 pKa = 5.83ATMYY349 pKa = 11.68NMFKK353 pKa = 9.83TLCLDD358 pKa = 3.66TASMLSPYY366 pKa = 9.18TFQRR370 pKa = 11.84EE371 pKa = 4.24FEE373 pKa = 4.37VEE375 pKa = 3.99PVAPAKK381 pKa = 10.26PWGEE385 pKa = 4.21CGDD388 pKa = 3.7LTLRR392 pKa = 11.84LPRR395 pKa = 11.84LNEE398 pKa = 3.66LLQRR402 pKa = 11.84LAVRR406 pKa = 11.84KK407 pKa = 8.99EE408 pKa = 3.63VTADD412 pKa = 3.43VVIDD416 pKa = 3.59TVRR419 pKa = 11.84RR420 pKa = 11.84LAHH423 pKa = 5.31EE424 pKa = 4.54EE425 pKa = 3.73KK426 pKa = 8.48WTTEE430 pKa = 3.1IDD432 pKa = 3.45RR433 pKa = 11.84DD434 pKa = 4.04EE435 pKa = 4.72FEE437 pKa = 4.48LWLSRR442 pKa = 11.84VVTEE446 pKa = 4.3AGEE449 pKa = 4.09RR450 pKa = 11.84AQPLTLAYY458 pKa = 9.78TNGQCWTCGTFGKK471 pKa = 7.08THH473 pKa = 5.6RR474 pKa = 11.84HH475 pKa = 4.33EE476 pKa = 4.95CKK478 pKa = 9.4ACKK481 pKa = 9.55RR482 pKa = 11.84RR483 pKa = 11.84ARR485 pKa = 11.84NRR487 pKa = 11.84VPDD490 pKa = 4.11RR491 pKa = 11.84PLSDD495 pKa = 4.21AFATHH500 pKa = 6.35MGFFPLWSEE509 pKa = 4.3RR510 pKa = 11.84FMLPNMAFKK519 pKa = 10.7RR520 pKa = 11.84GTQVRR525 pKa = 11.84VGKK528 pKa = 10.42AGNRR532 pKa = 11.84VMTTRR537 pKa = 11.84AEE539 pKa = 4.09VNAWLSRR546 pKa = 11.84QRR548 pKa = 11.84VDD550 pKa = 2.94VSQRR554 pKa = 11.84GRR556 pKa = 11.84LCGPMFCNQVPKK568 pKa = 10.43CYY570 pKa = 9.92PKK572 pKa = 10.44GQAVALSAFLVRR584 pKa = 11.84LGGTRR589 pKa = 11.84PYY591 pKa = 10.54PDD593 pKa = 3.6TFPCQMCASDD603 pKa = 3.58GHH605 pKa = 5.7EE606 pKa = 4.2EE607 pKa = 4.23CTKK610 pKa = 10.64QCTPDD615 pKa = 3.47EE616 pKa = 4.45LVFAPNNVRR625 pKa = 11.84PEE627 pKa = 3.79VKK629 pKa = 10.07LAYY632 pKa = 9.95DD633 pKa = 3.66LLYY636 pKa = 10.4TVFSQYY642 pKa = 10.86VLYY645 pKa = 10.92GGLTSASCPLKK656 pKa = 10.53PEE658 pKa = 3.84SRR660 pKa = 11.84TEE662 pKa = 4.01FLSHH666 pKa = 7.04FAGEE670 pKa = 4.46KK671 pKa = 8.52LLKK674 pKa = 9.71MVEE677 pKa = 4.06ALAMINDD684 pKa = 3.71GWLEE688 pKa = 3.97NMDD691 pKa = 4.21DD692 pKa = 3.42EE693 pKa = 4.61AVRR696 pKa = 11.84VRR698 pKa = 11.84FTGFTKK704 pKa = 10.62AEE706 pKa = 3.7KK707 pKa = 10.49SYY709 pKa = 11.01DD710 pKa = 3.74FEE712 pKa = 4.47WTGDD716 pKa = 3.66CLILKK721 pKa = 10.22KK722 pKa = 10.28EE723 pKa = 3.98GKK725 pKa = 9.28PRR727 pKa = 11.84FICSPDD733 pKa = 3.4PVMLYY738 pKa = 10.05YY739 pKa = 10.86LGRR742 pKa = 11.84YY743 pKa = 5.89THH745 pKa = 6.55VQTKK749 pKa = 8.5WLSARR754 pKa = 11.84FLPCSRR760 pKa = 11.84MFYY763 pKa = 10.22AGCATPEE770 pKa = 4.48DD771 pKa = 3.92MNVWLNYY778 pKa = 8.27TAGDD782 pKa = 3.73AQYY785 pKa = 10.46MNTIVDD791 pKa = 4.42DD792 pKa = 4.29VSAMDD797 pKa = 3.75SSFTKK802 pKa = 10.24TLLDD806 pKa = 3.3FHH808 pKa = 6.69EE809 pKa = 5.68RR810 pKa = 11.84IRR812 pKa = 11.84DD813 pKa = 3.48LQFPHH818 pKa = 6.31MAAHH822 pKa = 6.41TKK824 pKa = 9.84AAFRR828 pKa = 11.84GEE830 pKa = 3.58EE831 pKa = 4.14RR832 pKa = 11.84FWVRR836 pKa = 11.84IGEE839 pKa = 3.83IRR841 pKa = 11.84AFVEE845 pKa = 4.1WVNASGVPDD854 pKa = 3.7TSYY857 pKa = 11.76KK858 pKa = 9.03NTAPCLPLRR867 pKa = 11.84VFAIVHH873 pKa = 6.78AIWGMIGQPFEE884 pKa = 4.2LTIEE888 pKa = 4.34RR889 pKa = 11.84YY890 pKa = 9.95YY891 pKa = 11.31LVLDD895 pKa = 4.86GIYY898 pKa = 10.17TSAAGDD904 pKa = 3.79DD905 pKa = 3.85GLTRR909 pKa = 11.84VPTHH913 pKa = 5.89MFGVDD918 pKa = 3.1TSSPQFKK925 pKa = 9.69MGYY928 pKa = 8.31CKK930 pKa = 10.26AWSFFGFNVKK940 pKa = 9.79VDD942 pKa = 3.6IVPPCRR948 pKa = 11.84WRR950 pKa = 11.84MATYY954 pKa = 10.06LAQRR958 pKa = 11.84PVWVGHH964 pKa = 5.75RR965 pKa = 11.84YY966 pKa = 8.37EE967 pKa = 4.4WAPEE971 pKa = 3.46PARR974 pKa = 11.84RR975 pKa = 11.84LRR977 pKa = 11.84GIFWQIDD984 pKa = 3.77CSLHH988 pKa = 5.63PIAWGRR994 pKa = 11.84GIATQLLQQAKK1005 pKa = 8.68ALPVVSHH1012 pKa = 5.55VCQWYY1017 pKa = 9.05LARR1020 pKa = 11.84TSGPAIISAATAASHH1035 pKa = 6.57EE1036 pKa = 4.18YY1037 pKa = 10.78SPFHH1041 pKa = 6.49GAVCSGDD1048 pKa = 3.15INEE1051 pKa = 4.66RR1052 pKa = 11.84GVSEE1056 pKa = 4.21FCVDD1060 pKa = 3.43YY1061 pKa = 10.93NIPRR1065 pKa = 11.84AEE1067 pKa = 4.15LDD1069 pKa = 3.4RR1070 pKa = 11.84FCRR1073 pKa = 11.84LLEE1076 pKa = 4.09LVPSVLVNLNSFVLDD1091 pKa = 4.21RR1092 pKa = 11.84IYY1094 pKa = 11.24AEE1096 pKa = 4.3EE1097 pKa = 4.06SS1098 pKa = 3.15
Molecular weight: 123.92 kDa
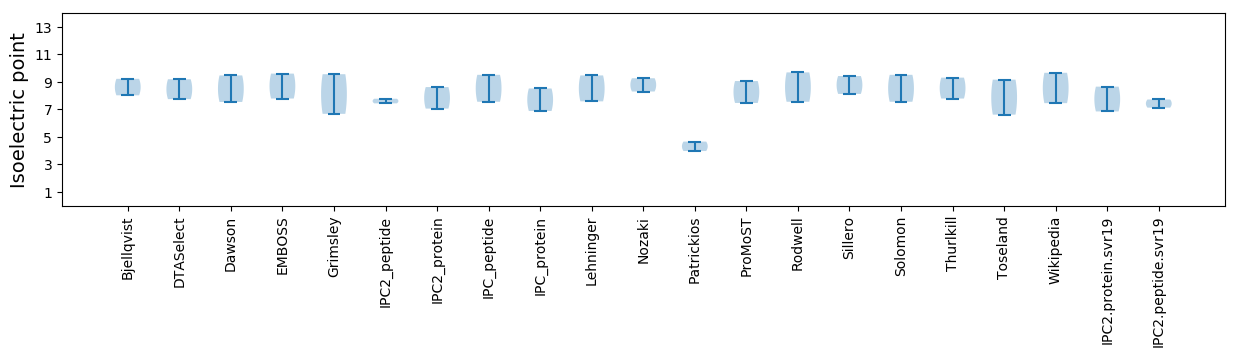
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KL89|A0A1L3KL89_9VIRU Uncharacterized protein OS=Shahe isopoda virus 3 OX=1923423 PE=4 SV=1
MM1 pKa = 7.73PSSKK5 pKa = 10.11PSQLDD10 pKa = 3.68LVSLPTLCKK19 pKa = 10.03TISLPGDD26 pKa = 3.69YY27 pKa = 10.5PPVRR31 pKa = 11.84LPTYY35 pKa = 9.07PAVEE39 pKa = 4.22RR40 pKa = 11.84TSVLPFNATLTTTVPSEE57 pKa = 4.07GMRR60 pKa = 11.84GMLIKK65 pKa = 10.67SSTMPLWTDD74 pKa = 3.4APVVAPYY81 pKa = 10.16SYY83 pKa = 11.31TMGWDD88 pKa = 3.29ISPTVGTPAATSSIWPIMPDD108 pKa = 2.94AKK110 pKa = 10.77GAGGGSFSADD120 pKa = 2.73VRR122 pKa = 11.84NGLEE126 pKa = 4.46VIGSNPPPGGYY137 pKa = 9.51PIVGEE142 pKa = 4.88LDD144 pKa = 3.33LKK146 pKa = 11.22AYY148 pKa = 9.76IYY150 pKa = 10.06LPKK153 pKa = 10.07GARR156 pKa = 11.84VLITAFGPSVPDD168 pKa = 3.11SGANLTLEE176 pKa = 4.07QAEE179 pKa = 4.34LGGTIASEE187 pKa = 4.06NYY189 pKa = 9.56ALNVGAGATASYY201 pKa = 10.3KK202 pKa = 10.54GYY204 pKa = 9.99FRR206 pKa = 11.84EE207 pKa = 4.08FTIPKK212 pKa = 9.89SFFARR217 pKa = 11.84PLSLSINRR225 pKa = 11.84AWYY228 pKa = 9.83GFTLNITVMNKK239 pKa = 9.05VPIMNASTGNMTLTWSPTEE258 pKa = 3.7PSIPLMVPAIKK269 pKa = 10.26PPALDD274 pKa = 3.1ISAIPFSNTRR284 pKa = 11.84VTALAALFTNVTKK297 pKa = 10.83ALNKK301 pKa = 9.89EE302 pKa = 4.17GTVMAGRR309 pKa = 11.84FNPATLDD316 pKa = 3.86ALDD319 pKa = 4.49LTMADD324 pKa = 3.89YY325 pKa = 11.03SQLAPCEE332 pKa = 4.21KK333 pKa = 10.58YY334 pKa = 10.55FFGLEE339 pKa = 3.94KK340 pKa = 10.74GFYY343 pKa = 9.81TYY345 pKa = 10.61IPLQTDD351 pKa = 3.12VGEE354 pKa = 4.45FADD357 pKa = 4.91DD358 pKa = 3.28VWDD361 pKa = 4.31APQISVTGGVRR372 pKa = 11.84IPWLHH377 pKa = 6.97LGRR380 pKa = 11.84TALVNTFRR388 pKa = 11.84FDD390 pKa = 3.93DD391 pKa = 4.52PDD393 pKa = 5.34GGTSLAVNLDD403 pKa = 2.98WHH405 pKa = 5.8VEE407 pKa = 3.94YY408 pKa = 10.85RR409 pKa = 11.84NSSVLWPVAVSALSLEE425 pKa = 4.42EE426 pKa = 3.61AHH428 pKa = 7.32RR429 pKa = 11.84AQLVCLEE436 pKa = 4.56AGFFFDD442 pKa = 5.4NVDD445 pKa = 3.27HH446 pKa = 6.98KK447 pKa = 11.09FILSMVSKK455 pKa = 10.13GLGMVAPMLKK465 pKa = 9.97NRR467 pKa = 11.84APAIASAGMAARR479 pKa = 11.84GYY481 pKa = 8.97IRR483 pKa = 11.84SYY485 pKa = 10.63LGEE488 pKa = 4.23SSHH491 pKa = 6.89PKK493 pKa = 9.76PSSLQVATAPRR504 pKa = 11.84VKK506 pKa = 10.21VRR508 pKa = 11.84HH509 pKa = 6.0PKK511 pKa = 10.1DD512 pKa = 3.5KK513 pKa = 10.29KK514 pKa = 10.84AKK516 pKa = 9.29AAPRR520 pKa = 11.84KK521 pKa = 9.35PKK523 pKa = 10.1SDD525 pKa = 3.03KK526 pKa = 10.55KK527 pKa = 10.65KK528 pKa = 10.4KK529 pKa = 10.19KK530 pKa = 9.88KK531 pKa = 10.33
MM1 pKa = 7.73PSSKK5 pKa = 10.11PSQLDD10 pKa = 3.68LVSLPTLCKK19 pKa = 10.03TISLPGDD26 pKa = 3.69YY27 pKa = 10.5PPVRR31 pKa = 11.84LPTYY35 pKa = 9.07PAVEE39 pKa = 4.22RR40 pKa = 11.84TSVLPFNATLTTTVPSEE57 pKa = 4.07GMRR60 pKa = 11.84GMLIKK65 pKa = 10.67SSTMPLWTDD74 pKa = 3.4APVVAPYY81 pKa = 10.16SYY83 pKa = 11.31TMGWDD88 pKa = 3.29ISPTVGTPAATSSIWPIMPDD108 pKa = 2.94AKK110 pKa = 10.77GAGGGSFSADD120 pKa = 2.73VRR122 pKa = 11.84NGLEE126 pKa = 4.46VIGSNPPPGGYY137 pKa = 9.51PIVGEE142 pKa = 4.88LDD144 pKa = 3.33LKK146 pKa = 11.22AYY148 pKa = 9.76IYY150 pKa = 10.06LPKK153 pKa = 10.07GARR156 pKa = 11.84VLITAFGPSVPDD168 pKa = 3.11SGANLTLEE176 pKa = 4.07QAEE179 pKa = 4.34LGGTIASEE187 pKa = 4.06NYY189 pKa = 9.56ALNVGAGATASYY201 pKa = 10.3KK202 pKa = 10.54GYY204 pKa = 9.99FRR206 pKa = 11.84EE207 pKa = 4.08FTIPKK212 pKa = 9.89SFFARR217 pKa = 11.84PLSLSINRR225 pKa = 11.84AWYY228 pKa = 9.83GFTLNITVMNKK239 pKa = 9.05VPIMNASTGNMTLTWSPTEE258 pKa = 3.7PSIPLMVPAIKK269 pKa = 10.26PPALDD274 pKa = 3.1ISAIPFSNTRR284 pKa = 11.84VTALAALFTNVTKK297 pKa = 10.83ALNKK301 pKa = 9.89EE302 pKa = 4.17GTVMAGRR309 pKa = 11.84FNPATLDD316 pKa = 3.86ALDD319 pKa = 4.49LTMADD324 pKa = 3.89YY325 pKa = 11.03SQLAPCEE332 pKa = 4.21KK333 pKa = 10.58YY334 pKa = 10.55FFGLEE339 pKa = 3.94KK340 pKa = 10.74GFYY343 pKa = 9.81TYY345 pKa = 10.61IPLQTDD351 pKa = 3.12VGEE354 pKa = 4.45FADD357 pKa = 4.91DD358 pKa = 3.28VWDD361 pKa = 4.31APQISVTGGVRR372 pKa = 11.84IPWLHH377 pKa = 6.97LGRR380 pKa = 11.84TALVNTFRR388 pKa = 11.84FDD390 pKa = 3.93DD391 pKa = 4.52PDD393 pKa = 5.34GGTSLAVNLDD403 pKa = 2.98WHH405 pKa = 5.8VEE407 pKa = 3.94YY408 pKa = 10.85RR409 pKa = 11.84NSSVLWPVAVSALSLEE425 pKa = 4.42EE426 pKa = 3.61AHH428 pKa = 7.32RR429 pKa = 11.84AQLVCLEE436 pKa = 4.56AGFFFDD442 pKa = 5.4NVDD445 pKa = 3.27HH446 pKa = 6.98KK447 pKa = 11.09FILSMVSKK455 pKa = 10.13GLGMVAPMLKK465 pKa = 9.97NRR467 pKa = 11.84APAIASAGMAARR479 pKa = 11.84GYY481 pKa = 8.97IRR483 pKa = 11.84SYY485 pKa = 10.63LGEE488 pKa = 4.23SSHH491 pKa = 6.89PKK493 pKa = 9.76PSSLQVATAPRR504 pKa = 11.84VKK506 pKa = 10.21VRR508 pKa = 11.84HH509 pKa = 6.0PKK511 pKa = 10.1DD512 pKa = 3.5KK513 pKa = 10.29KK514 pKa = 10.84AKK516 pKa = 9.29AAPRR520 pKa = 11.84KK521 pKa = 9.35PKK523 pKa = 10.1SDD525 pKa = 3.03KK526 pKa = 10.55KK527 pKa = 10.65KK528 pKa = 10.4KK529 pKa = 10.19KK530 pKa = 9.88KK531 pKa = 10.33
Molecular weight: 57.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1629 |
531 |
1098 |
814.5 |
90.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.699 ± 0.342 | 2.394 ± 0.95 |
4.604 ± 0.044 | 4.85 ± 0.758 |
4.911 ± 0.497 | 6.446 ± 0.565 |
1.78 ± 0.338 | 3.683 ± 0.434 |
4.174 ± 0.864 | 9.208 ± 0.01 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.885 ± 0.066 | 3.376 ± 0.203 |
7.612 ± 0.839 | 2.578 ± 0.654 |
6.2 ± 1.166 | 5.893 ± 1.243 |
6.507 ± 0.435 | 7.551 ± 0.303 |
2.087 ± 0.204 | 3.56 ± 0.089 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
