
Hirsutella minnesotensis 3608
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Ophiocordycipitaceae; Hirsutella; Hirsutella minnesotensis
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
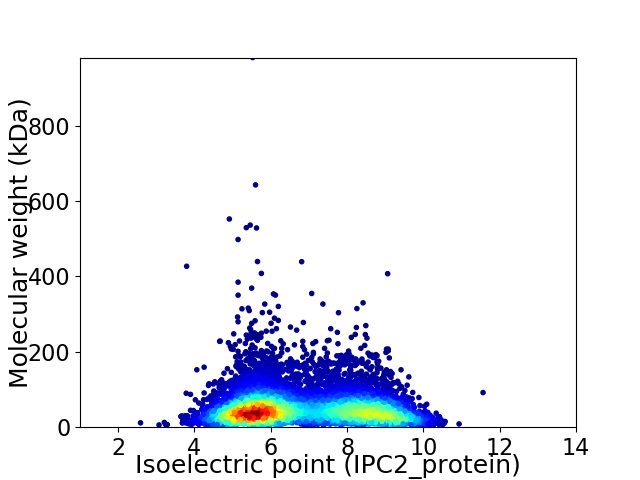
Virtual 2D-PAGE plot for 9072 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F7ZKY1|A0A0F7ZKY1_9HYPO Uncharacterized protein OS=Hirsutella minnesotensis 3608 OX=1043627 GN=HIM_04918 PE=4 SV=1
MM1 pKa = 7.6SNPDD5 pKa = 3.23LSMSAFSGSIAQDD18 pKa = 2.97DD19 pKa = 4.11VMRR22 pKa = 11.84FSSQAQEE29 pKa = 4.2GTASLSGDD37 pKa = 3.42LRR39 pKa = 11.84STRR42 pKa = 11.84CQILSPSCRR51 pKa = 11.84LIASAHH57 pKa = 4.4QSGGRR62 pKa = 11.84EE63 pKa = 3.95NHH65 pKa = 6.05SPPLPSTPEE74 pKa = 3.67PRR76 pKa = 11.84NVYY79 pKa = 9.34HH80 pKa = 6.32QPQHH84 pKa = 3.6THH86 pKa = 5.18YY87 pKa = 10.34RR88 pKa = 11.84PFDD91 pKa = 3.76APEE94 pKa = 3.99PSFDD98 pKa = 5.11DD99 pKa = 3.08SDD101 pKa = 4.35YY102 pKa = 11.38PQGLLQDD109 pKa = 3.65EE110 pKa = 5.04DD111 pKa = 4.13EE112 pKa = 5.12LVDD115 pKa = 4.21EE116 pKa = 5.78KK117 pKa = 11.13IRR119 pKa = 11.84DD120 pKa = 3.66TNFAYY125 pKa = 9.61IDD127 pKa = 3.92LAVCDD132 pKa = 4.51EE133 pKa = 5.08GYY135 pKa = 8.42STANDD140 pKa = 3.33QGSPTVSHH148 pKa = 6.81RR149 pKa = 11.84SVEE152 pKa = 4.35EE153 pKa = 3.66TCMDD157 pKa = 3.88CTCIRR162 pKa = 11.84EE163 pKa = 4.21ILNQSLDD170 pKa = 4.15DD171 pKa = 4.44FPLSTSEE178 pKa = 4.1LSPAGFMDD186 pKa = 5.01LCDD189 pKa = 3.94EE190 pKa = 4.79SPDD193 pKa = 3.79FGNPYY198 pKa = 10.52DD199 pKa = 5.12DD200 pKa = 3.96EE201 pKa = 5.73LSFIDD206 pKa = 5.37LTDD209 pKa = 3.46EE210 pKa = 4.29TDD212 pKa = 3.62DD213 pKa = 5.02DD214 pKa = 4.25EE215 pKa = 6.82LMVIDD220 pKa = 4.57LTEE223 pKa = 4.78DD224 pKa = 2.89SWSNTSANEE233 pKa = 3.84EE234 pKa = 4.21SRR236 pKa = 11.84GEE238 pKa = 3.98NPIQVISDD246 pKa = 3.34FVIVGEE252 pKa = 4.41SLCPVYY258 pKa = 10.26KK259 pKa = 10.41YY260 pKa = 10.47HH261 pKa = 7.16GVDD264 pKa = 5.64FIDD267 pKa = 3.81LTLLL271 pKa = 3.75
MM1 pKa = 7.6SNPDD5 pKa = 3.23LSMSAFSGSIAQDD18 pKa = 2.97DD19 pKa = 4.11VMRR22 pKa = 11.84FSSQAQEE29 pKa = 4.2GTASLSGDD37 pKa = 3.42LRR39 pKa = 11.84STRR42 pKa = 11.84CQILSPSCRR51 pKa = 11.84LIASAHH57 pKa = 4.4QSGGRR62 pKa = 11.84EE63 pKa = 3.95NHH65 pKa = 6.05SPPLPSTPEE74 pKa = 3.67PRR76 pKa = 11.84NVYY79 pKa = 9.34HH80 pKa = 6.32QPQHH84 pKa = 3.6THH86 pKa = 5.18YY87 pKa = 10.34RR88 pKa = 11.84PFDD91 pKa = 3.76APEE94 pKa = 3.99PSFDD98 pKa = 5.11DD99 pKa = 3.08SDD101 pKa = 4.35YY102 pKa = 11.38PQGLLQDD109 pKa = 3.65EE110 pKa = 5.04DD111 pKa = 4.13EE112 pKa = 5.12LVDD115 pKa = 4.21EE116 pKa = 5.78KK117 pKa = 11.13IRR119 pKa = 11.84DD120 pKa = 3.66TNFAYY125 pKa = 9.61IDD127 pKa = 3.92LAVCDD132 pKa = 4.51EE133 pKa = 5.08GYY135 pKa = 8.42STANDD140 pKa = 3.33QGSPTVSHH148 pKa = 6.81RR149 pKa = 11.84SVEE152 pKa = 4.35EE153 pKa = 3.66TCMDD157 pKa = 3.88CTCIRR162 pKa = 11.84EE163 pKa = 4.21ILNQSLDD170 pKa = 4.15DD171 pKa = 4.44FPLSTSEE178 pKa = 4.1LSPAGFMDD186 pKa = 5.01LCDD189 pKa = 3.94EE190 pKa = 4.79SPDD193 pKa = 3.79FGNPYY198 pKa = 10.52DD199 pKa = 5.12DD200 pKa = 3.96EE201 pKa = 5.73LSFIDD206 pKa = 5.37LTDD209 pKa = 3.46EE210 pKa = 4.29TDD212 pKa = 3.62DD213 pKa = 5.02DD214 pKa = 4.25EE215 pKa = 6.82LMVIDD220 pKa = 4.57LTEE223 pKa = 4.78DD224 pKa = 2.89SWSNTSANEE233 pKa = 3.84EE234 pKa = 4.21SRR236 pKa = 11.84GEE238 pKa = 3.98NPIQVISDD246 pKa = 3.34FVIVGEE252 pKa = 4.41SLCPVYY258 pKa = 10.26KK259 pKa = 10.41YY260 pKa = 10.47HH261 pKa = 7.16GVDD264 pKa = 5.64FIDD267 pKa = 3.81LTLLL271 pKa = 3.75
Molecular weight: 30.04 kDa
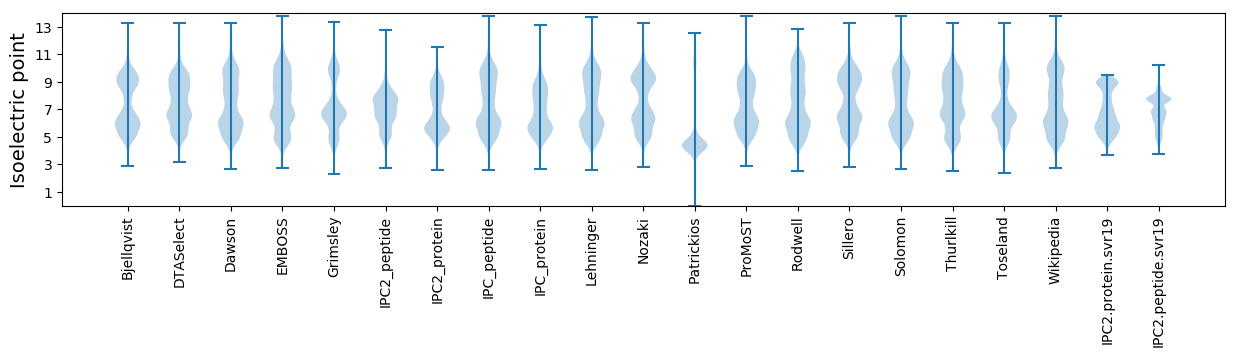
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F7ZIL0|A0A0F7ZIL0_9HYPO DDE_3 domain-containing protein OS=Hirsutella minnesotensis 3608 OX=1043627 GN=HIM_11931 PE=4 SV=1
MM1 pKa = 7.32YY2 pKa = 9.25PGLCWQTLQTRR13 pKa = 11.84DD14 pKa = 3.84KK15 pKa = 11.05KK16 pKa = 10.96RR17 pKa = 11.84LDD19 pKa = 3.58LGRR22 pKa = 11.84VDD24 pKa = 3.63PRR26 pKa = 11.84AAIGPSSPIVRR37 pKa = 11.84RR38 pKa = 11.84GGTTRR43 pKa = 11.84TTTLPRR49 pKa = 11.84AAIGPSSPIVRR60 pKa = 11.84RR61 pKa = 11.84GGTTRR66 pKa = 11.84TTTLPRR72 pKa = 11.84AAIGPSSPIVRR83 pKa = 11.84RR84 pKa = 11.84GGTTRR89 pKa = 11.84TTTLPRR95 pKa = 11.84AAIGPSSPIVRR106 pKa = 11.84RR107 pKa = 11.84GGTTRR112 pKa = 11.84TTTLPRR118 pKa = 11.84AAIGPSSPIVRR129 pKa = 11.84RR130 pKa = 11.84GGTTRR135 pKa = 11.84TTTLPRR141 pKa = 11.84AAIGPSSPIVRR152 pKa = 11.84RR153 pKa = 11.84GGTTRR158 pKa = 11.84TTTLPRR164 pKa = 11.84AAIGPSSPIVRR175 pKa = 11.84RR176 pKa = 11.84GGTTRR181 pKa = 11.84TTTLPRR187 pKa = 11.84AAIGPSSPIVRR198 pKa = 11.84RR199 pKa = 11.84GGTTRR204 pKa = 11.84TTTLPRR210 pKa = 11.84AAIGPSSPIVRR221 pKa = 11.84RR222 pKa = 11.84GGTTRR227 pKa = 11.84TTTLPRR233 pKa = 11.84AAIGPSSPIVRR244 pKa = 11.84RR245 pKa = 11.84GGTTRR250 pKa = 11.84TTTLPRR256 pKa = 11.84AAIGPSSPIVRR267 pKa = 11.84RR268 pKa = 11.84GGTTRR273 pKa = 11.84TTTLPRR279 pKa = 11.84AAIGPSSPIVRR290 pKa = 11.84RR291 pKa = 11.84GGTTRR296 pKa = 11.84TTTLPRR302 pKa = 11.84AAIGPSSPIVRR313 pKa = 11.84RR314 pKa = 11.84GGTTRR319 pKa = 11.84TTTLPRR325 pKa = 11.84AAIGPSSPIVRR336 pKa = 11.84RR337 pKa = 11.84GGTTRR342 pKa = 11.84TTTLPRR348 pKa = 11.84AAIGPSSPIVRR359 pKa = 11.84RR360 pKa = 11.84GGTTRR365 pKa = 11.84TTTLPRR371 pKa = 11.84AAIGPSSPIVRR382 pKa = 11.84RR383 pKa = 11.84GGTTRR388 pKa = 11.84TTTLPRR394 pKa = 11.84AAIGPSSPIVRR405 pKa = 11.84RR406 pKa = 11.84GGTTRR411 pKa = 11.84TTTLPRR417 pKa = 11.84AAIGPSSPIVRR428 pKa = 11.84RR429 pKa = 11.84GGTTRR434 pKa = 11.84TTTLPRR440 pKa = 11.84AAIGPSSPIVRR451 pKa = 11.84RR452 pKa = 11.84GGTTRR457 pKa = 11.84TTTLPRR463 pKa = 11.84AAIGPSSPIVRR474 pKa = 11.84RR475 pKa = 11.84GGTTRR480 pKa = 11.84TTTLPRR486 pKa = 11.84AAIGPSSPIVRR497 pKa = 11.84RR498 pKa = 11.84GGTTRR503 pKa = 11.84TTTLPRR509 pKa = 11.84AAIGPSSPIVRR520 pKa = 11.84RR521 pKa = 11.84GGTTRR526 pKa = 11.84TTTLPRR532 pKa = 11.84AAIGPSSPIVRR543 pKa = 11.84RR544 pKa = 11.84GGTTRR549 pKa = 11.84TTTLPRR555 pKa = 11.84AAIGPSSPIVRR566 pKa = 11.84RR567 pKa = 11.84GGTTRR572 pKa = 11.84TTTLPRR578 pKa = 11.84AAIGPSSPIVRR589 pKa = 11.84RR590 pKa = 11.84GGTTRR595 pKa = 11.84TTTLPRR601 pKa = 11.84AAIGPSSPIVRR612 pKa = 11.84RR613 pKa = 11.84GGTTRR618 pKa = 11.84TTTLPRR624 pKa = 11.84AAIGPSSPIVRR635 pKa = 11.84RR636 pKa = 11.84GGTTRR641 pKa = 11.84TTTLPRR647 pKa = 11.84AAIGPSSPIVRR658 pKa = 11.84RR659 pKa = 11.84GGTTRR664 pKa = 11.84TTTLPRR670 pKa = 11.84AAIGPSSPIVRR681 pKa = 11.84RR682 pKa = 11.84GGTTRR687 pKa = 11.84TTTLPRR693 pKa = 11.84AAIGPSSPIVRR704 pKa = 11.84RR705 pKa = 11.84GGTTRR710 pKa = 11.84TTTLPRR716 pKa = 11.84AAIGPSSPIVRR727 pKa = 11.84RR728 pKa = 11.84GGTTRR733 pKa = 11.84TTTLPRR739 pKa = 11.84AAIGPSSPIVRR750 pKa = 11.84RR751 pKa = 11.84GGTTRR756 pKa = 11.84TTTLPRR762 pKa = 11.84AAIGPSSPIVRR773 pKa = 11.84RR774 pKa = 11.84GGTTRR779 pKa = 11.84TTTLPRR785 pKa = 11.84AAIGPSSPIVRR796 pKa = 11.84RR797 pKa = 11.84GGTTRR802 pKa = 11.84TTTLPRR808 pKa = 11.84AAIGPSSPIVRR819 pKa = 11.84RR820 pKa = 11.84GGTTRR825 pKa = 11.84TTTLPRR831 pKa = 11.84AAIGPSSPIVRR842 pKa = 11.84RR843 pKa = 11.84GGTTRR848 pKa = 11.84TTTLPRR854 pKa = 11.84AAIGPSSPIVRR865 pKa = 11.84RR866 pKa = 11.84GGTTRR871 pKa = 11.84TTTLPRR877 pKa = 11.84AATVAPWKK885 pKa = 10.2WPINPDD891 pKa = 3.11KK892 pKa = 11.36GFF894 pKa = 3.53
MM1 pKa = 7.32YY2 pKa = 9.25PGLCWQTLQTRR13 pKa = 11.84DD14 pKa = 3.84KK15 pKa = 11.05KK16 pKa = 10.96RR17 pKa = 11.84LDD19 pKa = 3.58LGRR22 pKa = 11.84VDD24 pKa = 3.63PRR26 pKa = 11.84AAIGPSSPIVRR37 pKa = 11.84RR38 pKa = 11.84GGTTRR43 pKa = 11.84TTTLPRR49 pKa = 11.84AAIGPSSPIVRR60 pKa = 11.84RR61 pKa = 11.84GGTTRR66 pKa = 11.84TTTLPRR72 pKa = 11.84AAIGPSSPIVRR83 pKa = 11.84RR84 pKa = 11.84GGTTRR89 pKa = 11.84TTTLPRR95 pKa = 11.84AAIGPSSPIVRR106 pKa = 11.84RR107 pKa = 11.84GGTTRR112 pKa = 11.84TTTLPRR118 pKa = 11.84AAIGPSSPIVRR129 pKa = 11.84RR130 pKa = 11.84GGTTRR135 pKa = 11.84TTTLPRR141 pKa = 11.84AAIGPSSPIVRR152 pKa = 11.84RR153 pKa = 11.84GGTTRR158 pKa = 11.84TTTLPRR164 pKa = 11.84AAIGPSSPIVRR175 pKa = 11.84RR176 pKa = 11.84GGTTRR181 pKa = 11.84TTTLPRR187 pKa = 11.84AAIGPSSPIVRR198 pKa = 11.84RR199 pKa = 11.84GGTTRR204 pKa = 11.84TTTLPRR210 pKa = 11.84AAIGPSSPIVRR221 pKa = 11.84RR222 pKa = 11.84GGTTRR227 pKa = 11.84TTTLPRR233 pKa = 11.84AAIGPSSPIVRR244 pKa = 11.84RR245 pKa = 11.84GGTTRR250 pKa = 11.84TTTLPRR256 pKa = 11.84AAIGPSSPIVRR267 pKa = 11.84RR268 pKa = 11.84GGTTRR273 pKa = 11.84TTTLPRR279 pKa = 11.84AAIGPSSPIVRR290 pKa = 11.84RR291 pKa = 11.84GGTTRR296 pKa = 11.84TTTLPRR302 pKa = 11.84AAIGPSSPIVRR313 pKa = 11.84RR314 pKa = 11.84GGTTRR319 pKa = 11.84TTTLPRR325 pKa = 11.84AAIGPSSPIVRR336 pKa = 11.84RR337 pKa = 11.84GGTTRR342 pKa = 11.84TTTLPRR348 pKa = 11.84AAIGPSSPIVRR359 pKa = 11.84RR360 pKa = 11.84GGTTRR365 pKa = 11.84TTTLPRR371 pKa = 11.84AAIGPSSPIVRR382 pKa = 11.84RR383 pKa = 11.84GGTTRR388 pKa = 11.84TTTLPRR394 pKa = 11.84AAIGPSSPIVRR405 pKa = 11.84RR406 pKa = 11.84GGTTRR411 pKa = 11.84TTTLPRR417 pKa = 11.84AAIGPSSPIVRR428 pKa = 11.84RR429 pKa = 11.84GGTTRR434 pKa = 11.84TTTLPRR440 pKa = 11.84AAIGPSSPIVRR451 pKa = 11.84RR452 pKa = 11.84GGTTRR457 pKa = 11.84TTTLPRR463 pKa = 11.84AAIGPSSPIVRR474 pKa = 11.84RR475 pKa = 11.84GGTTRR480 pKa = 11.84TTTLPRR486 pKa = 11.84AAIGPSSPIVRR497 pKa = 11.84RR498 pKa = 11.84GGTTRR503 pKa = 11.84TTTLPRR509 pKa = 11.84AAIGPSSPIVRR520 pKa = 11.84RR521 pKa = 11.84GGTTRR526 pKa = 11.84TTTLPRR532 pKa = 11.84AAIGPSSPIVRR543 pKa = 11.84RR544 pKa = 11.84GGTTRR549 pKa = 11.84TTTLPRR555 pKa = 11.84AAIGPSSPIVRR566 pKa = 11.84RR567 pKa = 11.84GGTTRR572 pKa = 11.84TTTLPRR578 pKa = 11.84AAIGPSSPIVRR589 pKa = 11.84RR590 pKa = 11.84GGTTRR595 pKa = 11.84TTTLPRR601 pKa = 11.84AAIGPSSPIVRR612 pKa = 11.84RR613 pKa = 11.84GGTTRR618 pKa = 11.84TTTLPRR624 pKa = 11.84AAIGPSSPIVRR635 pKa = 11.84RR636 pKa = 11.84GGTTRR641 pKa = 11.84TTTLPRR647 pKa = 11.84AAIGPSSPIVRR658 pKa = 11.84RR659 pKa = 11.84GGTTRR664 pKa = 11.84TTTLPRR670 pKa = 11.84AAIGPSSPIVRR681 pKa = 11.84RR682 pKa = 11.84GGTTRR687 pKa = 11.84TTTLPRR693 pKa = 11.84AAIGPSSPIVRR704 pKa = 11.84RR705 pKa = 11.84GGTTRR710 pKa = 11.84TTTLPRR716 pKa = 11.84AAIGPSSPIVRR727 pKa = 11.84RR728 pKa = 11.84GGTTRR733 pKa = 11.84TTTLPRR739 pKa = 11.84AAIGPSSPIVRR750 pKa = 11.84RR751 pKa = 11.84GGTTRR756 pKa = 11.84TTTLPRR762 pKa = 11.84AAIGPSSPIVRR773 pKa = 11.84RR774 pKa = 11.84GGTTRR779 pKa = 11.84TTTLPRR785 pKa = 11.84AAIGPSSPIVRR796 pKa = 11.84RR797 pKa = 11.84GGTTRR802 pKa = 11.84TTTLPRR808 pKa = 11.84AAIGPSSPIVRR819 pKa = 11.84RR820 pKa = 11.84GGTTRR825 pKa = 11.84TTTLPRR831 pKa = 11.84AAIGPSSPIVRR842 pKa = 11.84RR843 pKa = 11.84GGTTRR848 pKa = 11.84TTTLPRR854 pKa = 11.84AAIGPSSPIVRR865 pKa = 11.84RR866 pKa = 11.84GGTTRR871 pKa = 11.84TTTLPRR877 pKa = 11.84AATVAPWKK885 pKa = 10.2WPINPDD891 pKa = 3.11KK892 pKa = 11.36GFF894 pKa = 3.53
Molecular weight: 91.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4418322 |
50 |
8878 |
487.0 |
54.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.181 ± 0.028 | 1.4 ± 0.009 |
5.923 ± 0.019 | 6.005 ± 0.03 |
3.6 ± 0.014 | 6.812 ± 0.028 |
2.52 ± 0.011 | 4.466 ± 0.016 |
4.641 ± 0.02 | 9.001 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.155 ± 0.011 | 3.333 ± 0.014 |
5.993 ± 0.027 | 4.234 ± 0.019 |
7.106 ± 0.025 | 7.869 ± 0.033 |
5.56 ± 0.015 | 6.124 ± 0.02 |
1.54 ± 0.009 | 2.538 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
