
Tico virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Tico phebovirus
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
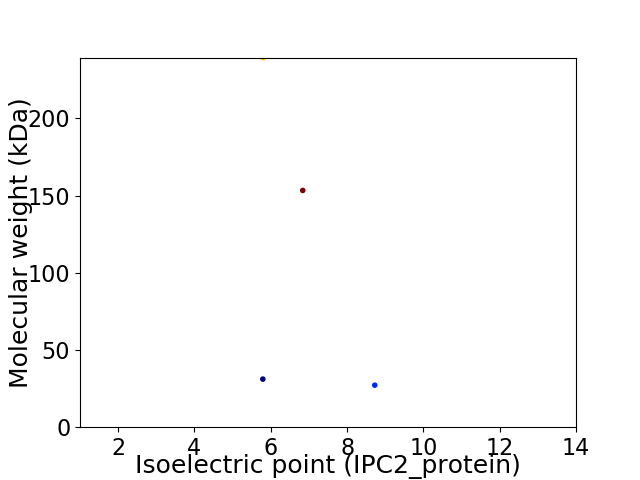
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
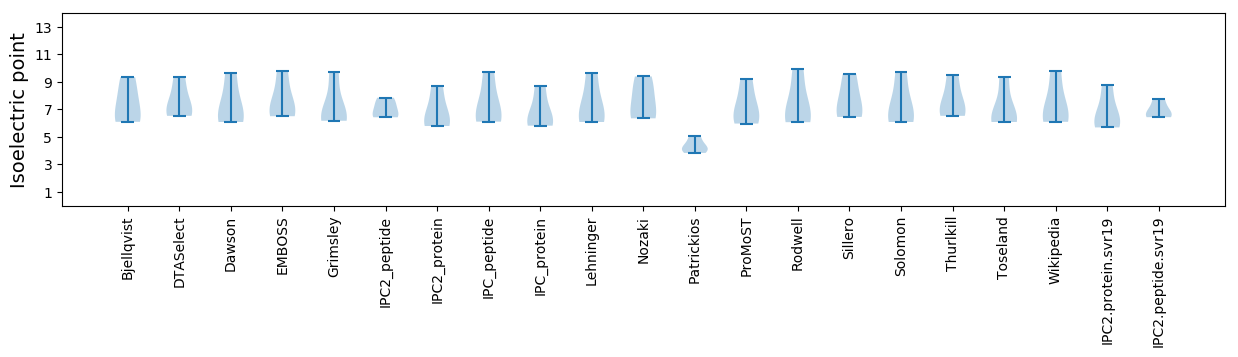
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A482KEP2|A0A482KEP2_9VIRU Nonstructural protein OS=Tico virus OX=2559113 GN=NSs PE=4 SV=1
MM1 pKa = 7.33ASKK4 pKa = 10.9YY5 pKa = 10.73LFDD8 pKa = 4.55RR9 pKa = 11.84PVVSRR14 pKa = 11.84TSDD17 pKa = 3.01QLKK20 pKa = 10.32RR21 pKa = 11.84IFVHH25 pKa = 4.96YY26 pKa = 8.95TAFNKK31 pKa = 8.88VCKK34 pKa = 10.48YY35 pKa = 9.95PITRR39 pKa = 11.84FMSLEE44 pKa = 4.11VPTVYY49 pKa = 10.69YY50 pKa = 10.7AVVEE54 pKa = 4.3DD55 pKa = 4.86EE56 pKa = 5.05RR57 pKa = 11.84PSLPDD62 pKa = 4.15FYY64 pKa = 11.65RR65 pKa = 11.84MGQLPARR72 pKa = 11.84WGFPEE77 pKa = 4.87DD78 pKa = 3.41SQVTKK83 pKa = 10.72RR84 pKa = 11.84SPRR87 pKa = 11.84KK88 pKa = 9.75YY89 pKa = 9.85LGLSIGLSSLDD100 pKa = 3.22SKK102 pKa = 11.56DD103 pKa = 3.31YY104 pKa = 11.31VRR106 pKa = 11.84FDD108 pKa = 3.52EE109 pKa = 5.1PLLKK113 pKa = 10.55SAISWPLTYY122 pKa = 9.82PSHH125 pKa = 6.8VFLNLLVKK133 pKa = 10.27RR134 pKa = 11.84EE135 pKa = 4.11SMGPWMFKK143 pKa = 10.26SICATYY149 pKa = 9.25FCRR152 pKa = 11.84MTNCEE157 pKa = 4.58EE158 pKa = 3.74IDD160 pKa = 3.41IALVRR165 pKa = 11.84AHH167 pKa = 7.23KK168 pKa = 10.32IIINEE173 pKa = 4.02ATKK176 pKa = 10.04MGLDD180 pKa = 3.08ITQFSGEE187 pKa = 4.1NLLMEE192 pKa = 4.84ICHH195 pKa = 5.61IQCIKK200 pKa = 10.65LLEE203 pKa = 4.42AFSNEE208 pKa = 3.61PSEE211 pKa = 4.54GRR213 pKa = 11.84VHH215 pKa = 7.24SPIYY219 pKa = 9.83QLLRR223 pKa = 11.84MFSNDD228 pKa = 2.31IVVPSLRR235 pKa = 11.84SGSGDD240 pKa = 3.43GDD242 pKa = 3.78CPALVNEE249 pKa = 4.98CVTAFIDD256 pKa = 5.31GMNGDD261 pKa = 4.01INHH264 pKa = 7.25PSVVSQDD271 pKa = 3.58YY272 pKa = 9.8EE273 pKa = 3.9
MM1 pKa = 7.33ASKK4 pKa = 10.9YY5 pKa = 10.73LFDD8 pKa = 4.55RR9 pKa = 11.84PVVSRR14 pKa = 11.84TSDD17 pKa = 3.01QLKK20 pKa = 10.32RR21 pKa = 11.84IFVHH25 pKa = 4.96YY26 pKa = 8.95TAFNKK31 pKa = 8.88VCKK34 pKa = 10.48YY35 pKa = 9.95PITRR39 pKa = 11.84FMSLEE44 pKa = 4.11VPTVYY49 pKa = 10.69YY50 pKa = 10.7AVVEE54 pKa = 4.3DD55 pKa = 4.86EE56 pKa = 5.05RR57 pKa = 11.84PSLPDD62 pKa = 4.15FYY64 pKa = 11.65RR65 pKa = 11.84MGQLPARR72 pKa = 11.84WGFPEE77 pKa = 4.87DD78 pKa = 3.41SQVTKK83 pKa = 10.72RR84 pKa = 11.84SPRR87 pKa = 11.84KK88 pKa = 9.75YY89 pKa = 9.85LGLSIGLSSLDD100 pKa = 3.22SKK102 pKa = 11.56DD103 pKa = 3.31YY104 pKa = 11.31VRR106 pKa = 11.84FDD108 pKa = 3.52EE109 pKa = 5.1PLLKK113 pKa = 10.55SAISWPLTYY122 pKa = 9.82PSHH125 pKa = 6.8VFLNLLVKK133 pKa = 10.27RR134 pKa = 11.84EE135 pKa = 4.11SMGPWMFKK143 pKa = 10.26SICATYY149 pKa = 9.25FCRR152 pKa = 11.84MTNCEE157 pKa = 4.58EE158 pKa = 3.74IDD160 pKa = 3.41IALVRR165 pKa = 11.84AHH167 pKa = 7.23KK168 pKa = 10.32IIINEE173 pKa = 4.02ATKK176 pKa = 10.04MGLDD180 pKa = 3.08ITQFSGEE187 pKa = 4.1NLLMEE192 pKa = 4.84ICHH195 pKa = 5.61IQCIKK200 pKa = 10.65LLEE203 pKa = 4.42AFSNEE208 pKa = 3.61PSEE211 pKa = 4.54GRR213 pKa = 11.84VHH215 pKa = 7.24SPIYY219 pKa = 9.83QLLRR223 pKa = 11.84MFSNDD228 pKa = 2.31IVVPSLRR235 pKa = 11.84SGSGDD240 pKa = 3.43GDD242 pKa = 3.78CPALVNEE249 pKa = 4.98CVTAFIDD256 pKa = 5.31GMNGDD261 pKa = 4.01INHH264 pKa = 7.25PSVVSQDD271 pKa = 3.58YY272 pKa = 9.8EE273 pKa = 3.9
Molecular weight: 31.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A482KC17|A0A482KC17_9VIRU Glycoprotein OS=Tico virus OX=2559113 GN=GPC PE=4 SV=1
MM1 pKa = 8.01SDD3 pKa = 3.61YY4 pKa = 11.2QKK6 pKa = 11.07LAVQFASEE14 pKa = 4.68DD15 pKa = 3.78LDD17 pKa = 3.62HH18 pKa = 6.99TAIQQWVNEE27 pKa = 4.04FAYY30 pKa = 10.13EE31 pKa = 4.12GFDD34 pKa = 3.08ARR36 pKa = 11.84TVIKK40 pKa = 10.47LLQEE44 pKa = 4.13RR45 pKa = 11.84GGDD48 pKa = 3.28GWKK51 pKa = 10.44NDD53 pKa = 3.63AKK55 pKa = 11.22KK56 pKa = 10.24MIVLALTRR64 pKa = 11.84GNKK67 pKa = 7.19PAKK70 pKa = 8.89MIKK73 pKa = 10.05KK74 pKa = 8.69MSPEE78 pKa = 3.53GRR80 pKa = 11.84KK81 pKa = 7.14TVEE84 pKa = 3.96ALVSKK89 pKa = 10.95YY90 pKa = 10.72KK91 pKa = 9.7LTSGNPSRR99 pKa = 11.84NDD101 pKa = 3.14LTLSRR106 pKa = 11.84VAAALAGWTCQAAVIVQDD124 pKa = 3.78YY125 pKa = 11.0LPVTGKK131 pKa = 9.05TIDD134 pKa = 3.69AFSPNYY140 pKa = 8.5PRR142 pKa = 11.84TMMHH146 pKa = 6.96PSFAGLIDD154 pKa = 3.5PTLPRR159 pKa = 11.84DD160 pKa = 3.47ILEE163 pKa = 4.56TIINAHH169 pKa = 6.18SLFLYY174 pKa = 10.26NFSRR178 pKa = 11.84TINPSLRR185 pKa = 11.84GLSKK189 pKa = 11.4AEE191 pKa = 3.88VEE193 pKa = 4.35QSFGQPMRR201 pKa = 11.84AAINSTFISSAQRR214 pKa = 11.84RR215 pKa = 11.84LMLEE219 pKa = 3.76TLGIMDD225 pKa = 4.74QNMKK229 pKa = 10.31LAANVVKK236 pKa = 10.52AAAIYY241 pKa = 9.75EE242 pKa = 4.05AMYY245 pKa = 11.01
MM1 pKa = 8.01SDD3 pKa = 3.61YY4 pKa = 11.2QKK6 pKa = 11.07LAVQFASEE14 pKa = 4.68DD15 pKa = 3.78LDD17 pKa = 3.62HH18 pKa = 6.99TAIQQWVNEE27 pKa = 4.04FAYY30 pKa = 10.13EE31 pKa = 4.12GFDD34 pKa = 3.08ARR36 pKa = 11.84TVIKK40 pKa = 10.47LLQEE44 pKa = 4.13RR45 pKa = 11.84GGDD48 pKa = 3.28GWKK51 pKa = 10.44NDD53 pKa = 3.63AKK55 pKa = 11.22KK56 pKa = 10.24MIVLALTRR64 pKa = 11.84GNKK67 pKa = 7.19PAKK70 pKa = 8.89MIKK73 pKa = 10.05KK74 pKa = 8.69MSPEE78 pKa = 3.53GRR80 pKa = 11.84KK81 pKa = 7.14TVEE84 pKa = 3.96ALVSKK89 pKa = 10.95YY90 pKa = 10.72KK91 pKa = 9.7LTSGNPSRR99 pKa = 11.84NDD101 pKa = 3.14LTLSRR106 pKa = 11.84VAAALAGWTCQAAVIVQDD124 pKa = 3.78YY125 pKa = 11.0LPVTGKK131 pKa = 9.05TIDD134 pKa = 3.69AFSPNYY140 pKa = 8.5PRR142 pKa = 11.84TMMHH146 pKa = 6.96PSFAGLIDD154 pKa = 3.5PTLPRR159 pKa = 11.84DD160 pKa = 3.47ILEE163 pKa = 4.56TIINAHH169 pKa = 6.18SLFLYY174 pKa = 10.26NFSRR178 pKa = 11.84TINPSLRR185 pKa = 11.84GLSKK189 pKa = 11.4AEE191 pKa = 3.88VEE193 pKa = 4.35QSFGQPMRR201 pKa = 11.84AAINSTFISSAQRR214 pKa = 11.84RR215 pKa = 11.84LMLEE219 pKa = 3.76TLGIMDD225 pKa = 4.74QNMKK229 pKa = 10.31LAANVVKK236 pKa = 10.52AAAIYY241 pKa = 9.75EE242 pKa = 4.05AMYY245 pKa = 11.01
Molecular weight: 27.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4000 |
245 |
2101 |
1000.0 |
112.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.35 ± 0.877 | 2.625 ± 0.786 |
5.55 ± 0.603 | 6.625 ± 0.449 |
4.5 ± 0.318 | 5.775 ± 0.252 |
2.675 ± 0.272 | 6.1 ± 0.377 |
5.95 ± 0.257 | 8.975 ± 0.257 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.475 ± 0.389 | 4.0 ± 0.258 |
3.725 ± 0.508 | 3.475 ± 0.298 |
5.7 ± 0.151 | 10.125 ± 0.74 |
4.975 ± 0.239 | 6.55 ± 0.205 |
1.175 ± 0.08 | 2.675 ± 0.405 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
