
Human genital-associated circular DNA virus-1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemygorvirus; Gemygorvirus sewopo1; Sewage derived gemygorvirus 1
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
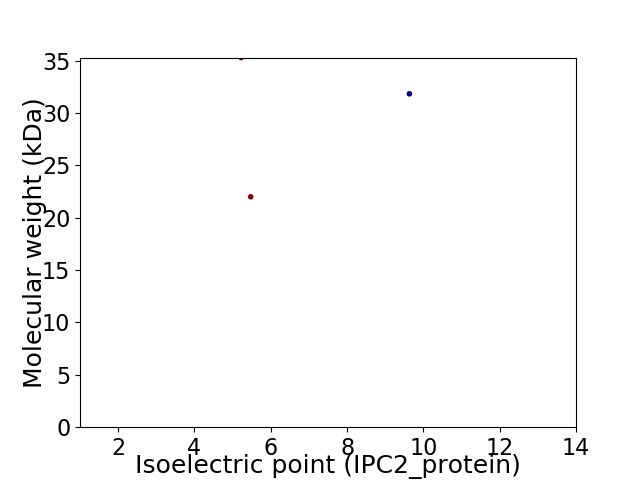
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E3DBM9|A0A0E3DBM9_9VIRU Capsid protein OS=Human genital-associated circular DNA virus-1 OX=1488574 PE=4 SV=1
MM1 pKa = 7.68AEE3 pKa = 4.03SPFRR7 pKa = 11.84VQSRR11 pKa = 11.84YY12 pKa = 9.12VLITYY17 pKa = 7.44AQCGEE22 pKa = 4.12LDD24 pKa = 3.3PWAVHH29 pKa = 7.07DD30 pKa = 5.8SITSYY35 pKa = 10.23PAEE38 pKa = 3.97CLIGRR43 pKa = 11.84EE44 pKa = 3.92NHH46 pKa = 6.51ADD48 pKa = 3.55GGIHH52 pKa = 6.42LHH54 pKa = 6.48AFVDD58 pKa = 4.58FGRR61 pKa = 11.84RR62 pKa = 11.84INLRR66 pKa = 11.84DD67 pKa = 3.48PRR69 pKa = 11.84RR70 pKa = 11.84FDD72 pKa = 3.43VDD74 pKa = 3.69DD75 pKa = 3.85RR76 pKa = 11.84HH77 pKa = 6.96PNIQPCGRR85 pKa = 11.84TPQKK89 pKa = 10.33MLDD92 pKa = 3.61YY93 pKa = 10.33AIKK96 pKa = 10.91DD97 pKa = 3.49GDD99 pKa = 4.03VVAGGLSTRR108 pKa = 11.84IGTEE112 pKa = 3.27IQGTDD117 pKa = 3.56NVWSRR122 pKa = 11.84IADD125 pKa = 3.58AASVEE130 pKa = 4.45EE131 pKa = 4.24FWEE134 pKa = 4.42LVRR137 pKa = 11.84EE138 pKa = 4.31LAPRR142 pKa = 11.84ALLCNHH148 pKa = 5.94QSLRR152 pKa = 11.84AYY154 pKa = 10.25AEE156 pKa = 3.52WHH158 pKa = 5.46YY159 pKa = 10.89RR160 pKa = 11.84PPAVAYY166 pKa = 6.77EE167 pKa = 4.33HH168 pKa = 7.2PPGLQLCTGKK178 pKa = 10.46FPEE181 pKa = 4.44LDD183 pKa = 2.66AWVRR187 pKa = 11.84DD188 pKa = 3.56NLLILWGEE196 pKa = 4.02TRR198 pKa = 11.84LGKK201 pKa = 7.89TVWARR206 pKa = 11.84SLGPHH211 pKa = 5.97IHH213 pKa = 7.3CCLQWNVDD221 pKa = 3.84DD222 pKa = 5.15LKK224 pKa = 11.47SGLEE228 pKa = 3.87DD229 pKa = 2.87AKK231 pKa = 11.26YY232 pKa = 10.73AVLDD236 pKa = 4.77DD237 pKa = 3.68IQGNFQFFPAYY248 pKa = 9.6KK249 pKa = 9.57GWLGGQSTFTVTDD262 pKa = 3.89KK263 pKa = 11.39YY264 pKa = 10.75RR265 pKa = 11.84GKK267 pKa = 7.59TTINWGRR274 pKa = 11.84PTIWLMNDD282 pKa = 3.12DD283 pKa = 4.37PEE285 pKa = 4.59EE286 pKa = 4.23VGHH289 pKa = 7.34VDD291 pKa = 4.77FNWLQGNCDD300 pKa = 3.97IIHH303 pKa = 6.02LTEE306 pKa = 4.03SLIVV310 pKa = 3.41
MM1 pKa = 7.68AEE3 pKa = 4.03SPFRR7 pKa = 11.84VQSRR11 pKa = 11.84YY12 pKa = 9.12VLITYY17 pKa = 7.44AQCGEE22 pKa = 4.12LDD24 pKa = 3.3PWAVHH29 pKa = 7.07DD30 pKa = 5.8SITSYY35 pKa = 10.23PAEE38 pKa = 3.97CLIGRR43 pKa = 11.84EE44 pKa = 3.92NHH46 pKa = 6.51ADD48 pKa = 3.55GGIHH52 pKa = 6.42LHH54 pKa = 6.48AFVDD58 pKa = 4.58FGRR61 pKa = 11.84RR62 pKa = 11.84INLRR66 pKa = 11.84DD67 pKa = 3.48PRR69 pKa = 11.84RR70 pKa = 11.84FDD72 pKa = 3.43VDD74 pKa = 3.69DD75 pKa = 3.85RR76 pKa = 11.84HH77 pKa = 6.96PNIQPCGRR85 pKa = 11.84TPQKK89 pKa = 10.33MLDD92 pKa = 3.61YY93 pKa = 10.33AIKK96 pKa = 10.91DD97 pKa = 3.49GDD99 pKa = 4.03VVAGGLSTRR108 pKa = 11.84IGTEE112 pKa = 3.27IQGTDD117 pKa = 3.56NVWSRR122 pKa = 11.84IADD125 pKa = 3.58AASVEE130 pKa = 4.45EE131 pKa = 4.24FWEE134 pKa = 4.42LVRR137 pKa = 11.84EE138 pKa = 4.31LAPRR142 pKa = 11.84ALLCNHH148 pKa = 5.94QSLRR152 pKa = 11.84AYY154 pKa = 10.25AEE156 pKa = 3.52WHH158 pKa = 5.46YY159 pKa = 10.89RR160 pKa = 11.84PPAVAYY166 pKa = 6.77EE167 pKa = 4.33HH168 pKa = 7.2PPGLQLCTGKK178 pKa = 10.46FPEE181 pKa = 4.44LDD183 pKa = 2.66AWVRR187 pKa = 11.84DD188 pKa = 3.56NLLILWGEE196 pKa = 4.02TRR198 pKa = 11.84LGKK201 pKa = 7.89TVWARR206 pKa = 11.84SLGPHH211 pKa = 5.97IHH213 pKa = 7.3CCLQWNVDD221 pKa = 3.84DD222 pKa = 5.15LKK224 pKa = 11.47SGLEE228 pKa = 3.87DD229 pKa = 2.87AKK231 pKa = 11.26YY232 pKa = 10.73AVLDD236 pKa = 4.77DD237 pKa = 3.68IQGNFQFFPAYY248 pKa = 9.6KK249 pKa = 9.57GWLGGQSTFTVTDD262 pKa = 3.89KK263 pKa = 11.39YY264 pKa = 10.75RR265 pKa = 11.84GKK267 pKa = 7.59TTINWGRR274 pKa = 11.84PTIWLMNDD282 pKa = 3.12DD283 pKa = 4.37PEE285 pKa = 4.59EE286 pKa = 4.23VGHH289 pKa = 7.34VDD291 pKa = 4.77FNWLQGNCDD300 pKa = 3.97IIHH303 pKa = 6.02LTEE306 pKa = 4.03SLIVV310 pKa = 3.41
Molecular weight: 35.29 kDa
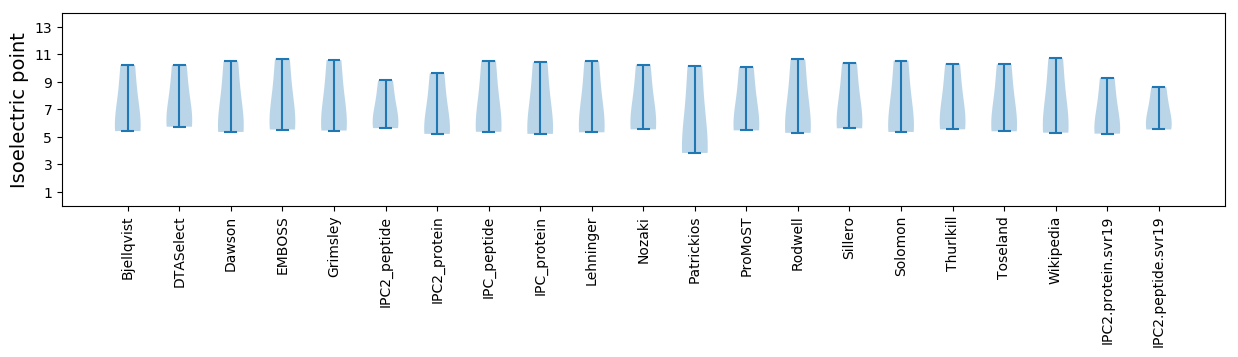
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3DBM9|A0A0E3DBM9_9VIRU Capsid protein OS=Human genital-associated circular DNA virus-1 OX=1488574 PE=4 SV=1
MM1 pKa = 7.4PRR3 pKa = 11.84RR4 pKa = 11.84YY5 pKa = 7.91TRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84ARR13 pKa = 11.84TSRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84STFRR22 pKa = 11.84KK23 pKa = 7.52TVTTRR28 pKa = 11.84GRR30 pKa = 11.84RR31 pKa = 11.84TRR33 pKa = 11.84KK34 pKa = 9.79YY35 pKa = 9.63SGARR39 pKa = 11.84NSRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84ISNITSIKK52 pKa = 9.97KK53 pKa = 9.2HH54 pKa = 6.5DD55 pKa = 3.62NMMPGFISPDD65 pKa = 2.82GTFNLNPLTTTTGFTSPFCPSARR88 pKa = 11.84RR89 pKa = 11.84LGLHH93 pKa = 6.85AEE95 pKa = 4.5GEE97 pKa = 4.66SARR100 pKa = 11.84QRR102 pKa = 11.84QTTFSVGYY110 pKa = 9.58KK111 pKa = 9.78EE112 pKa = 4.64RR113 pKa = 11.84LQIDD117 pKa = 3.77VLGDD121 pKa = 3.95GVWKK125 pKa = 9.08WRR127 pKa = 11.84RR128 pKa = 11.84VVFTYY133 pKa = 10.15KK134 pKa = 10.46GRR136 pKa = 11.84SLYY139 pKa = 10.86DD140 pKa = 3.27QDD142 pKa = 4.27TPWIVPYY149 pKa = 9.85YY150 pKa = 10.32NKK152 pKa = 9.93VVDD155 pKa = 4.11PTGHH159 pKa = 5.75GVDD162 pKa = 3.5MVRR165 pKa = 11.84AIAQPTLDD173 pKa = 3.35QAAHH177 pKa = 6.18IRR179 pKa = 11.84SVWWDD184 pKa = 3.37GFEE187 pKa = 4.25GGDD190 pKa = 3.11WGSEE194 pKa = 3.85FTAKK198 pKa = 10.5LDD200 pKa = 3.63TSRR203 pKa = 11.84ITPMYY208 pKa = 10.59DD209 pKa = 2.43RR210 pKa = 11.84TVTLNPGDD218 pKa = 3.87EE219 pKa = 4.48SGMSRR224 pKa = 11.84TFRR227 pKa = 11.84QWHH230 pKa = 5.8PARR233 pKa = 11.84KK234 pKa = 9.31NLVYY238 pKa = 10.8DD239 pKa = 4.54DD240 pKa = 6.05DD241 pKa = 5.44EE242 pKa = 7.64DD243 pKa = 4.03DD244 pKa = 3.79TAPTDD249 pKa = 3.52GGSPVSVTGKK259 pKa = 9.77PGMGDD264 pKa = 3.73LYY266 pKa = 11.33VYY268 pKa = 10.13DD269 pKa = 3.37IVYY272 pKa = 10.52NAVPATGG279 pKa = 3.29
MM1 pKa = 7.4PRR3 pKa = 11.84RR4 pKa = 11.84YY5 pKa = 7.91TRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84ARR13 pKa = 11.84TSRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84STFRR22 pKa = 11.84KK23 pKa = 7.52TVTTRR28 pKa = 11.84GRR30 pKa = 11.84RR31 pKa = 11.84TRR33 pKa = 11.84KK34 pKa = 9.79YY35 pKa = 9.63SGARR39 pKa = 11.84NSRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84ISNITSIKK52 pKa = 9.97KK53 pKa = 9.2HH54 pKa = 6.5DD55 pKa = 3.62NMMPGFISPDD65 pKa = 2.82GTFNLNPLTTTTGFTSPFCPSARR88 pKa = 11.84RR89 pKa = 11.84LGLHH93 pKa = 6.85AEE95 pKa = 4.5GEE97 pKa = 4.66SARR100 pKa = 11.84QRR102 pKa = 11.84QTTFSVGYY110 pKa = 9.58KK111 pKa = 9.78EE112 pKa = 4.64RR113 pKa = 11.84LQIDD117 pKa = 3.77VLGDD121 pKa = 3.95GVWKK125 pKa = 9.08WRR127 pKa = 11.84RR128 pKa = 11.84VVFTYY133 pKa = 10.15KK134 pKa = 10.46GRR136 pKa = 11.84SLYY139 pKa = 10.86DD140 pKa = 3.27QDD142 pKa = 4.27TPWIVPYY149 pKa = 9.85YY150 pKa = 10.32NKK152 pKa = 9.93VVDD155 pKa = 4.11PTGHH159 pKa = 5.75GVDD162 pKa = 3.5MVRR165 pKa = 11.84AIAQPTLDD173 pKa = 3.35QAAHH177 pKa = 6.18IRR179 pKa = 11.84SVWWDD184 pKa = 3.37GFEE187 pKa = 4.25GGDD190 pKa = 3.11WGSEE194 pKa = 3.85FTAKK198 pKa = 10.5LDD200 pKa = 3.63TSRR203 pKa = 11.84ITPMYY208 pKa = 10.59DD209 pKa = 2.43RR210 pKa = 11.84TVTLNPGDD218 pKa = 3.87EE219 pKa = 4.48SGMSRR224 pKa = 11.84TFRR227 pKa = 11.84QWHH230 pKa = 5.8PARR233 pKa = 11.84KK234 pKa = 9.31NLVYY238 pKa = 10.8DD239 pKa = 4.54DD240 pKa = 6.05DD241 pKa = 5.44EE242 pKa = 7.64DD243 pKa = 4.03DD244 pKa = 3.79TAPTDD249 pKa = 3.52GGSPVSVTGKK259 pKa = 9.77PGMGDD264 pKa = 3.73LYY266 pKa = 11.33VYY268 pKa = 10.13DD269 pKa = 3.37IVYY272 pKa = 10.52NAVPATGG279 pKa = 3.29
Molecular weight: 31.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
784 |
195 |
310 |
261.3 |
29.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.888 ± 0.96 | 1.786 ± 0.619 |
7.908 ± 0.085 | 4.592 ± 0.921 |
3.444 ± 0.125 | 8.291 ± 0.305 |
3.189 ± 0.61 | 4.847 ± 0.563 |
3.061 ± 0.435 | 7.27 ± 1.304 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.531 ± 0.424 | 3.444 ± 0.201 |
5.995 ± 0.27 | 3.316 ± 0.353 |
9.311 ± 1.454 | 5.102 ± 0.753 |
6.888 ± 1.702 | 6.378 ± 0.187 |
3.061 ± 0.374 | 3.699 ± 0.271 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
