
Beihai tombus-like virus 10
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.29
Get precalculated fractions of proteins
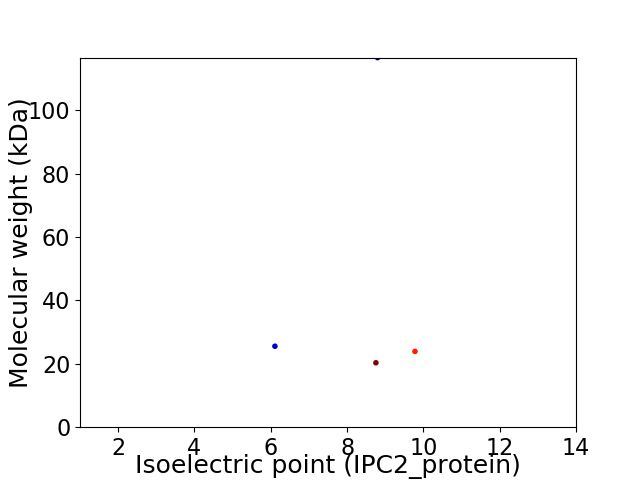
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFX5|A0A1L3KFX5_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 10 OX=1922713 PE=4 SV=1
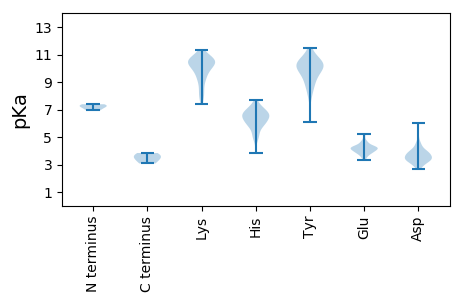
MM1 pKa = 6.94KK2 pKa = 9.84TVTINTKK9 pKa = 7.58WTADD13 pKa = 3.85SPDD16 pKa = 3.15SWLLNKK22 pKa = 9.92PYY24 pKa = 10.4RR25 pKa = 11.84IDD27 pKa = 3.49YY28 pKa = 8.45TEE30 pKa = 5.22KK31 pKa = 8.7IHH33 pKa = 7.02SVTVPALEE41 pKa = 4.26PEE43 pKa = 3.85ISIDD47 pKa = 3.43NVTTAEE53 pKa = 4.03YY54 pKa = 9.79MVGDD58 pKa = 4.13SSTNTMWPDD67 pKa = 3.2GPEE70 pKa = 3.45RR71 pKa = 11.84DD72 pKa = 3.89KK73 pKa = 11.18IYY75 pKa = 10.62VRR77 pKa = 11.84HH78 pKa = 6.11SFLGSINATQWLKK91 pKa = 9.49TATIVACYY99 pKa = 9.77SLSVKK104 pKa = 10.54YY105 pKa = 10.44KK106 pKa = 9.5ILQNGEE112 pKa = 4.07PNRR115 pKa = 11.84NGAGVFGRR123 pKa = 11.84DD124 pKa = 3.21LQLFRR129 pKa = 11.84HH130 pKa = 6.76DD131 pKa = 4.06LTVMATSPKK140 pKa = 10.29GITTAVVCPLVANTVVNVLDD160 pKa = 4.29DD161 pKa = 4.32RR162 pKa = 11.84VLQSLEE168 pKa = 3.98FKK170 pKa = 8.4TTFVVVAGWTYY181 pKa = 8.81TFVMHH186 pKa = 7.32SYY188 pKa = 8.92MRR190 pKa = 11.84PFFVNYY196 pKa = 9.29KK197 pKa = 9.69VEE199 pKa = 4.38AGTFIVFSAVATGAFMTYY217 pKa = 9.39IYY219 pKa = 10.11PGEE222 pKa = 4.26HH223 pKa = 5.14QVSLTT228 pKa = 3.42
MM1 pKa = 6.94KK2 pKa = 9.84TVTINTKK9 pKa = 7.58WTADD13 pKa = 3.85SPDD16 pKa = 3.15SWLLNKK22 pKa = 9.92PYY24 pKa = 10.4RR25 pKa = 11.84IDD27 pKa = 3.49YY28 pKa = 8.45TEE30 pKa = 5.22KK31 pKa = 8.7IHH33 pKa = 7.02SVTVPALEE41 pKa = 4.26PEE43 pKa = 3.85ISIDD47 pKa = 3.43NVTTAEE53 pKa = 4.03YY54 pKa = 9.79MVGDD58 pKa = 4.13SSTNTMWPDD67 pKa = 3.2GPEE70 pKa = 3.45RR71 pKa = 11.84DD72 pKa = 3.89KK73 pKa = 11.18IYY75 pKa = 10.62VRR77 pKa = 11.84HH78 pKa = 6.11SFLGSINATQWLKK91 pKa = 9.49TATIVACYY99 pKa = 9.77SLSVKK104 pKa = 10.54YY105 pKa = 10.44KK106 pKa = 9.5ILQNGEE112 pKa = 4.07PNRR115 pKa = 11.84NGAGVFGRR123 pKa = 11.84DD124 pKa = 3.21LQLFRR129 pKa = 11.84HH130 pKa = 6.76DD131 pKa = 4.06LTVMATSPKK140 pKa = 10.29GITTAVVCPLVANTVVNVLDD160 pKa = 4.29DD161 pKa = 4.32RR162 pKa = 11.84VLQSLEE168 pKa = 3.98FKK170 pKa = 8.4TTFVVVAGWTYY181 pKa = 8.81TFVMHH186 pKa = 7.32SYY188 pKa = 8.92MRR190 pKa = 11.84PFFVNYY196 pKa = 9.29KK197 pKa = 9.69VEE199 pKa = 4.38AGTFIVFSAVATGAFMTYY217 pKa = 9.39IYY219 pKa = 10.11PGEE222 pKa = 4.26HH223 pKa = 5.14QVSLTT228 pKa = 3.42
Molecular weight: 25.54 kDa
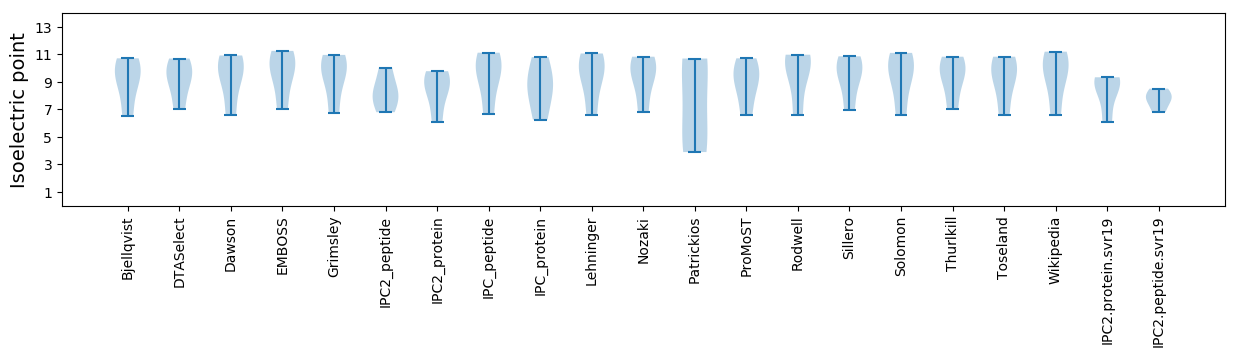
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFN7|A0A1L3KFN7_9VIRU RNA-directed RNA polymerase OS=Beihai tombus-like virus 10 OX=1922713 PE=4 SV=1
MM1 pKa = 7.38TGRR4 pKa = 11.84GRR6 pKa = 11.84GRR8 pKa = 11.84GRR10 pKa = 11.84GRR12 pKa = 11.84GRR14 pKa = 11.84GRR16 pKa = 11.84GGRR19 pKa = 11.84RR20 pKa = 11.84APPQPRR26 pKa = 11.84PPPPQPPPPQASGRR40 pKa = 11.84QLARR44 pKa = 11.84RR45 pKa = 11.84MARR48 pKa = 11.84MSIGRR53 pKa = 11.84QMNTGKK59 pKa = 8.1TQIRR63 pKa = 11.84RR64 pKa = 11.84NEE66 pKa = 3.76LCIADD71 pKa = 3.98VTSKK75 pKa = 10.59FATAKK80 pKa = 9.88RR81 pKa = 11.84FRR83 pKa = 11.84MCPGHH88 pKa = 6.23TGMMLLDD95 pKa = 3.92AMGTIHH101 pKa = 7.44SSYY104 pKa = 10.05KK105 pKa = 7.66YY106 pKa = 9.52HH107 pKa = 6.09SVKK110 pKa = 10.23IHH112 pKa = 4.74VQGMGPTTSKK122 pKa = 10.91CILHH126 pKa = 6.63ICADD130 pKa = 3.56ASVGAGAVTPTAVLQTIPNATCRR153 pKa = 11.84AWQTTTMNIPSTILNRR169 pKa = 11.84YY170 pKa = 6.06NHH172 pKa = 6.64YY173 pKa = 10.14ISHH176 pKa = 7.01EE177 pKa = 4.12ATASDD182 pKa = 3.37QAEE185 pKa = 4.61PFTLIIIGAGSADD198 pKa = 3.59GDD200 pKa = 3.74DD201 pKa = 3.39VTFLVKK207 pKa = 10.5VNYY210 pKa = 10.16NVTFFNPEE218 pKa = 3.77PQQ220 pKa = 3.08
MM1 pKa = 7.38TGRR4 pKa = 11.84GRR6 pKa = 11.84GRR8 pKa = 11.84GRR10 pKa = 11.84GRR12 pKa = 11.84GRR14 pKa = 11.84GRR16 pKa = 11.84GGRR19 pKa = 11.84RR20 pKa = 11.84APPQPRR26 pKa = 11.84PPPPQPPPPQASGRR40 pKa = 11.84QLARR44 pKa = 11.84RR45 pKa = 11.84MARR48 pKa = 11.84MSIGRR53 pKa = 11.84QMNTGKK59 pKa = 8.1TQIRR63 pKa = 11.84RR64 pKa = 11.84NEE66 pKa = 3.76LCIADD71 pKa = 3.98VTSKK75 pKa = 10.59FATAKK80 pKa = 9.88RR81 pKa = 11.84FRR83 pKa = 11.84MCPGHH88 pKa = 6.23TGMMLLDD95 pKa = 3.92AMGTIHH101 pKa = 7.44SSYY104 pKa = 10.05KK105 pKa = 7.66YY106 pKa = 9.52HH107 pKa = 6.09SVKK110 pKa = 10.23IHH112 pKa = 4.74VQGMGPTTSKK122 pKa = 10.91CILHH126 pKa = 6.63ICADD130 pKa = 3.56ASVGAGAVTPTAVLQTIPNATCRR153 pKa = 11.84AWQTTTMNIPSTILNRR169 pKa = 11.84YY170 pKa = 6.06NHH172 pKa = 6.64YY173 pKa = 10.14ISHH176 pKa = 7.01EE177 pKa = 4.12ATASDD182 pKa = 3.37QAEE185 pKa = 4.61PFTLIIIGAGSADD198 pKa = 3.59GDD200 pKa = 3.74DD201 pKa = 3.39VTFLVKK207 pKa = 10.5VNYY210 pKa = 10.16NVTFFNPEE218 pKa = 3.77PQQ220 pKa = 3.08
Molecular weight: 23.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1671 |
177 |
1046 |
417.8 |
46.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.54 ± 0.574 | 1.676 ± 0.231 |
4.728 ± 0.555 | 4.668 ± 0.751 |
3.291 ± 0.391 | 6.523 ± 0.901 |
3.052 ± 0.472 | 4.428 ± 0.57 |
6.882 ± 1.23 | 7.421 ± 1.11 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.232 ± 0.576 | 3.591 ± 0.552 |
6.942 ± 1.065 | 4.608 ± 1.405 |
6.403 ± 1.023 | 6.643 ± 0.611 |
6.942 ± 1.401 | 7.181 ± 1.252 |
0.958 ± 0.318 | 3.232 ± 0.66 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
