
Paenibacillus nanensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Paenibacillus
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
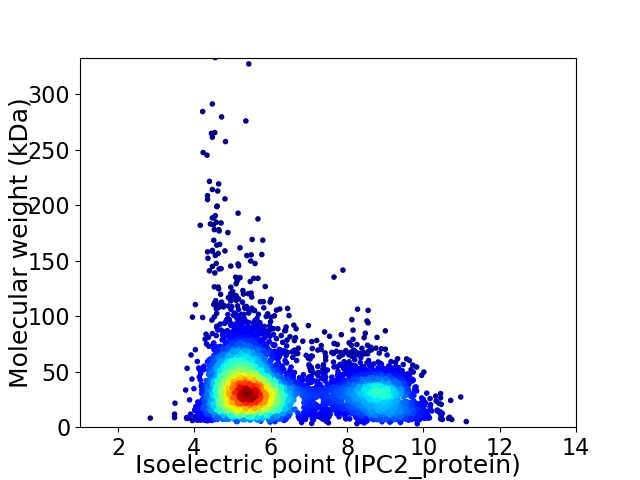
Virtual 2D-PAGE plot for 5258 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
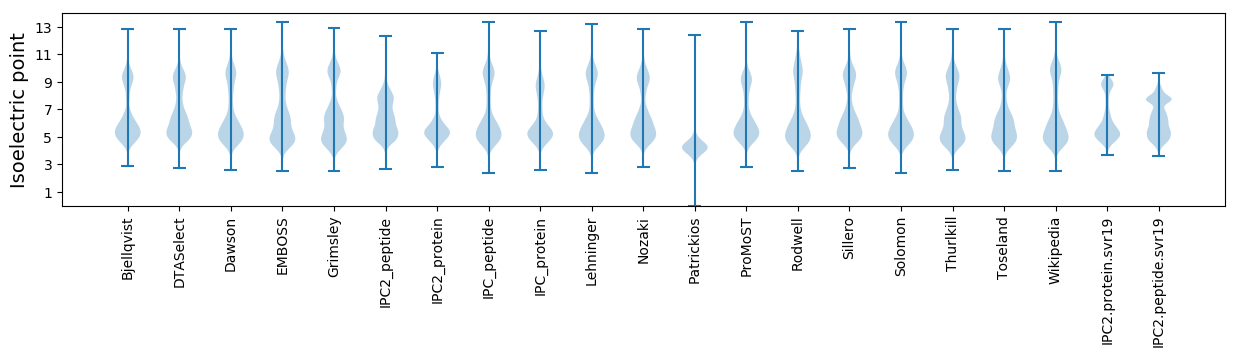
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A1UTI4|A0A3A1UTI4_9BACL Uncharacterized protein OS=Paenibacillus nanensis OX=393251 GN=D3P08_15390 PE=4 SV=1
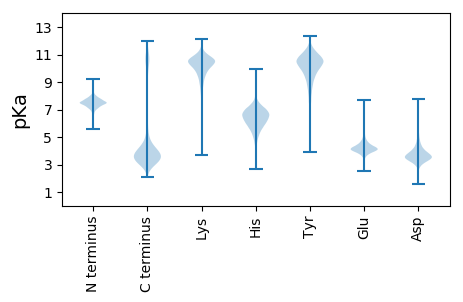
MM1 pKa = 7.95RR2 pKa = 11.84KK3 pKa = 7.21QTKK6 pKa = 9.59KK7 pKa = 10.09PFSIVMALLLTLTLTMSSFVYY28 pKa = 10.75AEE30 pKa = 3.99GLGDD34 pKa = 3.84EE35 pKa = 4.5EE36 pKa = 5.01KK37 pKa = 9.95KK38 pKa = 9.25TVGLQGEE45 pKa = 4.26PAGASLAGEE54 pKa = 4.03AHH56 pKa = 6.57AASDD60 pKa = 3.86EE61 pKa = 4.12QTEE64 pKa = 4.45ASCTTYY70 pKa = 11.05YY71 pKa = 11.33GFADD75 pKa = 4.49KK76 pKa = 10.6LAQQDD81 pKa = 3.52PTVPYY86 pKa = 9.22ITINGDD92 pKa = 3.05GTATTKK98 pKa = 10.66FKK100 pKa = 8.74TTQDD104 pKa = 3.33CVQASFSVYY113 pKa = 9.82TYY115 pKa = 11.06GEE117 pKa = 4.16SPTPYY122 pKa = 9.67EE123 pKa = 3.84NQVYY127 pKa = 10.17YY128 pKa = 10.77SGEE131 pKa = 3.73TAQYY135 pKa = 10.63DD136 pKa = 3.51KK137 pKa = 11.51AGNYY141 pKa = 7.98EE142 pKa = 4.21LTISIPVCGNSQIDD156 pKa = 4.49LYY158 pKa = 11.02GGPIQTTLGPGGHH171 pKa = 6.62KK172 pKa = 10.19PGTYY176 pKa = 9.3RR177 pKa = 11.84SYY179 pKa = 11.52LYY181 pKa = 9.91IIKK184 pKa = 10.02SPCPTPTPTDD194 pKa = 3.97PPTEE198 pKa = 4.41TPTTTPTDD206 pKa = 4.05PPTEE210 pKa = 4.41TPTTTPTDD218 pKa = 4.05PPTEE222 pKa = 4.41TPTTTPTDD230 pKa = 4.05PPTEE234 pKa = 4.41TPTTTPTDD242 pKa = 4.05PPTEE246 pKa = 4.39TPTTTPSDD254 pKa = 4.17PPTEE258 pKa = 4.47TPTTTPTDD266 pKa = 4.05PPTEE270 pKa = 4.41TPTTTPTDD278 pKa = 4.05PPTEE282 pKa = 4.39TPTTTPSDD290 pKa = 4.17PPTEE294 pKa = 4.47TPTTTPTDD302 pKa = 4.05PPTEE306 pKa = 4.41TPTTTPTDD314 pKa = 4.05PPTEE318 pKa = 4.39TPTTTPSDD326 pKa = 4.17PPTEE330 pKa = 4.45TPTTTPSDD338 pKa = 4.17PPTEE342 pKa = 4.45TPTTTPSDD350 pKa = 4.17PPTEE354 pKa = 4.47TPTTTPTQTPTSTPTQTPTSTPSWTPTPPPTWTWTPTPTPTPTTTPTATPTSTPTATPTSTPTDD418 pKa = 4.05PPTEE422 pKa = 4.47SPTPTPTDD430 pKa = 3.54TPTEE434 pKa = 4.36APSPTPTATTIITEE448 pKa = 4.24EE449 pKa = 4.31TPPDD453 pKa = 3.89DD454 pKa = 4.64GGTGGEE460 pKa = 4.08TDD462 pKa = 3.87GDD464 pKa = 3.88EE465 pKa = 4.21GSIPIDD471 pKa = 3.54SLPQTGDD478 pKa = 3.4SSPMPYY484 pKa = 10.42YY485 pKa = 11.19LLGTFIAVSGIVALRR500 pKa = 11.84KK501 pKa = 9.33RR502 pKa = 11.84RR503 pKa = 11.84RR504 pKa = 11.84QPP506 pKa = 2.87
MM1 pKa = 7.95RR2 pKa = 11.84KK3 pKa = 7.21QTKK6 pKa = 9.59KK7 pKa = 10.09PFSIVMALLLTLTLTMSSFVYY28 pKa = 10.75AEE30 pKa = 3.99GLGDD34 pKa = 3.84EE35 pKa = 4.5EE36 pKa = 5.01KK37 pKa = 9.95KK38 pKa = 9.25TVGLQGEE45 pKa = 4.26PAGASLAGEE54 pKa = 4.03AHH56 pKa = 6.57AASDD60 pKa = 3.86EE61 pKa = 4.12QTEE64 pKa = 4.45ASCTTYY70 pKa = 11.05YY71 pKa = 11.33GFADD75 pKa = 4.49KK76 pKa = 10.6LAQQDD81 pKa = 3.52PTVPYY86 pKa = 9.22ITINGDD92 pKa = 3.05GTATTKK98 pKa = 10.66FKK100 pKa = 8.74TTQDD104 pKa = 3.33CVQASFSVYY113 pKa = 9.82TYY115 pKa = 11.06GEE117 pKa = 4.16SPTPYY122 pKa = 9.67EE123 pKa = 3.84NQVYY127 pKa = 10.17YY128 pKa = 10.77SGEE131 pKa = 3.73TAQYY135 pKa = 10.63DD136 pKa = 3.51KK137 pKa = 11.51AGNYY141 pKa = 7.98EE142 pKa = 4.21LTISIPVCGNSQIDD156 pKa = 4.49LYY158 pKa = 11.02GGPIQTTLGPGGHH171 pKa = 6.62KK172 pKa = 10.19PGTYY176 pKa = 9.3RR177 pKa = 11.84SYY179 pKa = 11.52LYY181 pKa = 9.91IIKK184 pKa = 10.02SPCPTPTPTDD194 pKa = 3.97PPTEE198 pKa = 4.41TPTTTPTDD206 pKa = 4.05PPTEE210 pKa = 4.41TPTTTPTDD218 pKa = 4.05PPTEE222 pKa = 4.41TPTTTPTDD230 pKa = 4.05PPTEE234 pKa = 4.41TPTTTPTDD242 pKa = 4.05PPTEE246 pKa = 4.39TPTTTPSDD254 pKa = 4.17PPTEE258 pKa = 4.47TPTTTPTDD266 pKa = 4.05PPTEE270 pKa = 4.41TPTTTPTDD278 pKa = 4.05PPTEE282 pKa = 4.39TPTTTPSDD290 pKa = 4.17PPTEE294 pKa = 4.47TPTTTPTDD302 pKa = 4.05PPTEE306 pKa = 4.41TPTTTPTDD314 pKa = 4.05PPTEE318 pKa = 4.39TPTTTPSDD326 pKa = 4.17PPTEE330 pKa = 4.45TPTTTPSDD338 pKa = 4.17PPTEE342 pKa = 4.45TPTTTPSDD350 pKa = 4.17PPTEE354 pKa = 4.47TPTTTPTQTPTSTPTQTPTSTPSWTPTPPPTWTWTPTPTPTPTTTPTATPTSTPTATPTSTPTDD418 pKa = 4.05PPTEE422 pKa = 4.47SPTPTPTDD430 pKa = 3.54TPTEE434 pKa = 4.36APSPTPTATTIITEE448 pKa = 4.24EE449 pKa = 4.31TPPDD453 pKa = 3.89DD454 pKa = 4.64GGTGGEE460 pKa = 4.08TDD462 pKa = 3.87GDD464 pKa = 3.88EE465 pKa = 4.21GSIPIDD471 pKa = 3.54SLPQTGDD478 pKa = 3.4SSPMPYY484 pKa = 10.42YY485 pKa = 11.19LLGTFIAVSGIVALRR500 pKa = 11.84KK501 pKa = 9.33RR502 pKa = 11.84RR503 pKa = 11.84RR504 pKa = 11.84QPP506 pKa = 2.87
Molecular weight: 53.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A1URQ4|A0A3A1URQ4_9BACL DUF115 domain-containing protein OS=Paenibacillus nanensis OX=393251 GN=D3P08_25120 PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84PTFKK6 pKa = 10.84PNVSKK11 pKa = 10.73RR12 pKa = 11.84AKK14 pKa = 8.72VHH16 pKa = 5.43GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTKK25 pKa = 10.18NGRR28 pKa = 11.84KK29 pKa = 8.78VLAARR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.23GRR39 pKa = 11.84KK40 pKa = 8.56VLSAA44 pKa = 4.05
MM1 pKa = 7.9RR2 pKa = 11.84PTFKK6 pKa = 10.84PNVSKK11 pKa = 10.73RR12 pKa = 11.84AKK14 pKa = 8.72VHH16 pKa = 5.43GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTKK25 pKa = 10.18NGRR28 pKa = 11.84KK29 pKa = 8.78VLAARR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.23GRR39 pKa = 11.84KK40 pKa = 8.56VLSAA44 pKa = 4.05
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1792965 |
26 |
3082 |
341.0 |
37.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.076 ± 0.044 | 0.698 ± 0.011 |
5.159 ± 0.026 | 7.003 ± 0.038 |
4.09 ± 0.026 | 7.674 ± 0.031 |
1.999 ± 0.014 | 6.372 ± 0.031 |
5.067 ± 0.025 | 10.024 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.818 ± 0.017 | 3.642 ± 0.028 |
4.093 ± 0.021 | 3.566 ± 0.021 |
5.281 ± 0.037 | 6.286 ± 0.025 |
5.308 ± 0.033 | 6.982 ± 0.028 |
1.338 ± 0.015 | 3.523 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
