
Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Nostocaceae; Nostoc;
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
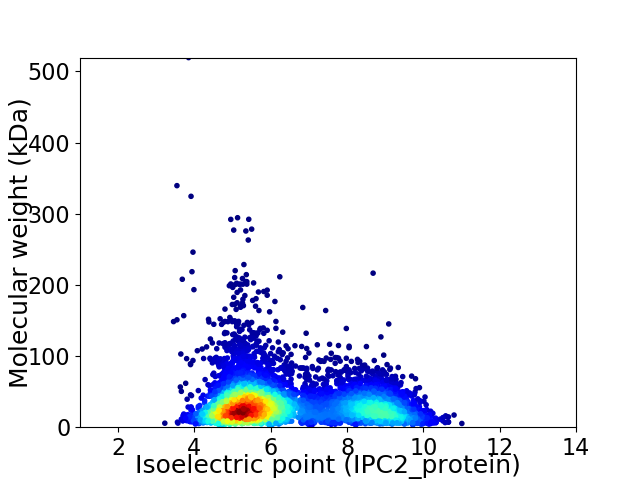
Virtual 2D-PAGE plot for 6071 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8YRM1|Q8YRM1_NOSS1 Alr3425 protein OS=Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) OX=103690 GN=alr3425 PE=4 SV=1
MM1 pKa = 6.93GTYY4 pKa = 10.47NGTTPFPDD12 pKa = 4.36PGSSTSTALDD22 pKa = 3.24TGLLIGGSLLSTPSLFQDD40 pKa = 3.22NLSRR44 pKa = 11.84LIPSDD49 pKa = 3.1LSDD52 pKa = 3.65YY53 pKa = 11.38YY54 pKa = 11.22RR55 pKa = 11.84FTLSSSSIVTLEE67 pKa = 4.73LDD69 pKa = 3.6GLTDD73 pKa = 3.66NVNLFLRR80 pKa = 11.84NSAGSSLAASSNAGTTAEE98 pKa = 4.69RR99 pKa = 11.84IRR101 pKa = 11.84FSISDD106 pKa = 3.63PPDD109 pKa = 3.24NPFYY113 pKa = 11.12AHH115 pKa = 7.26AFIPSGSISTPYY127 pKa = 10.66DD128 pKa = 3.18LRR130 pKa = 11.84LSAEE134 pKa = 4.44VIQQNNINDD143 pKa = 3.59NTIANARR150 pKa = 11.84NIGTFNTTTSITPVTDD166 pKa = 3.5YY167 pKa = 11.44VSGSGQVIDD176 pKa = 3.7QNDD179 pKa = 4.33YY180 pKa = 11.67YY181 pKa = 11.63SFTLTNSGTVDD192 pKa = 3.0INLNSLNGTDD202 pKa = 3.52TLYY205 pKa = 11.39ADD207 pKa = 4.34LQLISSSNSVIQTSATVGTSLEE229 pKa = 4.34SISRR233 pKa = 11.84SLAAGTYY240 pKa = 8.71YY241 pKa = 9.95IRR243 pKa = 11.84AYY245 pKa = 10.13SQSDD249 pKa = 3.24PGNYY253 pKa = 9.33NLEE256 pKa = 4.6FNFSADD262 pKa = 3.62PPDD265 pKa = 4.16AGGNTSDD272 pKa = 3.27TSTPINLPATFSEE285 pKa = 4.62IISDD289 pKa = 3.87QVSLGDD295 pKa = 3.65SSDD298 pKa = 3.97YY299 pKa = 11.15YY300 pKa = 11.43QFTLASASLVEE311 pKa = 4.18IQFTSLTADD320 pKa = 3.11ANLYY324 pKa = 10.34LQTQNGGNILSSTQPGTALDD344 pKa = 3.89AVRR347 pKa = 11.84LSLNAGTYY355 pKa = 10.33NILVNRR361 pKa = 11.84GSTEE365 pKa = 3.79TAQYY369 pKa = 9.0TLSGFAQAIGNDD381 pKa = 3.46QAPNSTTLALNLNLNSPISLNEE403 pKa = 3.54FVGNIDD409 pKa = 3.31TNDD412 pKa = 3.38YY413 pKa = 11.53YY414 pKa = 11.56KK415 pKa = 10.39FTVNGTTEE423 pKa = 3.86INLDD427 pKa = 3.64LSILNSYY434 pKa = 10.48LLDD437 pKa = 4.15PQLVNADD444 pKa = 3.69VQILNSGGTQVAISNQTGNSNEE466 pKa = 4.42SINTILGAGTYY477 pKa = 9.13FIRR480 pKa = 11.84VYY482 pKa = 10.62TSGLANTFYY491 pKa = 11.07DD492 pKa = 4.8LNITAQSQALLVQDD506 pKa = 4.55INPTGNSDD514 pKa = 3.7PANLTTLGNTLYY526 pKa = 9.07FTANDD531 pKa = 5.27GINGVQLWSSNGDD544 pKa = 2.67ITTRR548 pKa = 11.84LSNISSFNPTNLIVFNNRR566 pKa = 11.84LYY568 pKa = 10.66FAASNDD574 pKa = 3.55TFGRR578 pKa = 11.84EE579 pKa = 3.4LWEE582 pKa = 4.21YY583 pKa = 11.43NGTTVNRR590 pKa = 11.84ISDD593 pKa = 3.77INVGAGNSNPGNLTVAGNKK612 pKa = 10.0LFFTAVDD619 pKa = 3.32NDD621 pKa = 4.56SIRR624 pKa = 11.84KK625 pKa = 7.89LWVYY629 pKa = 11.14NGTNVNLVDD638 pKa = 4.0VNASFSNSSTPTFTTTFNNQLFFTAQNNSQLWSTDD673 pKa = 2.65GTIGGTQVISAGGITKK689 pKa = 8.0STPRR693 pKa = 11.84NLTVVGNTLYY703 pKa = 9.09FTANNGTSGHH713 pKa = 6.36EE714 pKa = 3.17IWQYY718 pKa = 11.25QNGTTASLLEE728 pKa = 5.61DD729 pKa = 3.28ITPGNNSFAPSSLTAVGNTLYY750 pKa = 10.7FVTDD754 pKa = 3.26SDD756 pKa = 3.98NDD758 pKa = 3.59FNLEE762 pKa = 3.89LWKK765 pKa = 10.77SDD767 pKa = 2.9GTAAGTDD774 pKa = 3.48IIGTDD779 pKa = 3.52GQAPNLGLGSFSLTAVDD796 pKa = 3.25NTLYY800 pKa = 10.55FVANDD805 pKa = 4.7PISGLEE811 pKa = 3.75LWKK814 pKa = 9.82TDD816 pKa = 3.05GTFANTAVVRR826 pKa = 11.84NISSGAADD834 pKa = 3.89SLPTGLVNFNGTLAFAASDD853 pKa = 4.42GINRR857 pKa = 11.84EE858 pKa = 3.93VWFSNGTEE866 pKa = 3.95LNTRR870 pKa = 11.84KK871 pKa = 9.98VSNINPTGNANPASLTVVGTKK892 pKa = 10.57LFFTANGANGTEE904 pKa = 4.5LYY906 pKa = 10.85VII908 pKa = 4.86
MM1 pKa = 6.93GTYY4 pKa = 10.47NGTTPFPDD12 pKa = 4.36PGSSTSTALDD22 pKa = 3.24TGLLIGGSLLSTPSLFQDD40 pKa = 3.22NLSRR44 pKa = 11.84LIPSDD49 pKa = 3.1LSDD52 pKa = 3.65YY53 pKa = 11.38YY54 pKa = 11.22RR55 pKa = 11.84FTLSSSSIVTLEE67 pKa = 4.73LDD69 pKa = 3.6GLTDD73 pKa = 3.66NVNLFLRR80 pKa = 11.84NSAGSSLAASSNAGTTAEE98 pKa = 4.69RR99 pKa = 11.84IRR101 pKa = 11.84FSISDD106 pKa = 3.63PPDD109 pKa = 3.24NPFYY113 pKa = 11.12AHH115 pKa = 7.26AFIPSGSISTPYY127 pKa = 10.66DD128 pKa = 3.18LRR130 pKa = 11.84LSAEE134 pKa = 4.44VIQQNNINDD143 pKa = 3.59NTIANARR150 pKa = 11.84NIGTFNTTTSITPVTDD166 pKa = 3.5YY167 pKa = 11.44VSGSGQVIDD176 pKa = 3.7QNDD179 pKa = 4.33YY180 pKa = 11.67YY181 pKa = 11.63SFTLTNSGTVDD192 pKa = 3.0INLNSLNGTDD202 pKa = 3.52TLYY205 pKa = 11.39ADD207 pKa = 4.34LQLISSSNSVIQTSATVGTSLEE229 pKa = 4.34SISRR233 pKa = 11.84SLAAGTYY240 pKa = 8.71YY241 pKa = 9.95IRR243 pKa = 11.84AYY245 pKa = 10.13SQSDD249 pKa = 3.24PGNYY253 pKa = 9.33NLEE256 pKa = 4.6FNFSADD262 pKa = 3.62PPDD265 pKa = 4.16AGGNTSDD272 pKa = 3.27TSTPINLPATFSEE285 pKa = 4.62IISDD289 pKa = 3.87QVSLGDD295 pKa = 3.65SSDD298 pKa = 3.97YY299 pKa = 11.15YY300 pKa = 11.43QFTLASASLVEE311 pKa = 4.18IQFTSLTADD320 pKa = 3.11ANLYY324 pKa = 10.34LQTQNGGNILSSTQPGTALDD344 pKa = 3.89AVRR347 pKa = 11.84LSLNAGTYY355 pKa = 10.33NILVNRR361 pKa = 11.84GSTEE365 pKa = 3.79TAQYY369 pKa = 9.0TLSGFAQAIGNDD381 pKa = 3.46QAPNSTTLALNLNLNSPISLNEE403 pKa = 3.54FVGNIDD409 pKa = 3.31TNDD412 pKa = 3.38YY413 pKa = 11.53YY414 pKa = 11.56KK415 pKa = 10.39FTVNGTTEE423 pKa = 3.86INLDD427 pKa = 3.64LSILNSYY434 pKa = 10.48LLDD437 pKa = 4.15PQLVNADD444 pKa = 3.69VQILNSGGTQVAISNQTGNSNEE466 pKa = 4.42SINTILGAGTYY477 pKa = 9.13FIRR480 pKa = 11.84VYY482 pKa = 10.62TSGLANTFYY491 pKa = 11.07DD492 pKa = 4.8LNITAQSQALLVQDD506 pKa = 4.55INPTGNSDD514 pKa = 3.7PANLTTLGNTLYY526 pKa = 9.07FTANDD531 pKa = 5.27GINGVQLWSSNGDD544 pKa = 2.67ITTRR548 pKa = 11.84LSNISSFNPTNLIVFNNRR566 pKa = 11.84LYY568 pKa = 10.66FAASNDD574 pKa = 3.55TFGRR578 pKa = 11.84EE579 pKa = 3.4LWEE582 pKa = 4.21YY583 pKa = 11.43NGTTVNRR590 pKa = 11.84ISDD593 pKa = 3.77INVGAGNSNPGNLTVAGNKK612 pKa = 10.0LFFTAVDD619 pKa = 3.32NDD621 pKa = 4.56SIRR624 pKa = 11.84KK625 pKa = 7.89LWVYY629 pKa = 11.14NGTNVNLVDD638 pKa = 4.0VNASFSNSSTPTFTTTFNNQLFFTAQNNSQLWSTDD673 pKa = 2.65GTIGGTQVISAGGITKK689 pKa = 8.0STPRR693 pKa = 11.84NLTVVGNTLYY703 pKa = 9.09FTANNGTSGHH713 pKa = 6.36EE714 pKa = 3.17IWQYY718 pKa = 11.25QNGTTASLLEE728 pKa = 5.61DD729 pKa = 3.28ITPGNNSFAPSSLTAVGNTLYY750 pKa = 10.7FVTDD754 pKa = 3.26SDD756 pKa = 3.98NDD758 pKa = 3.59FNLEE762 pKa = 3.89LWKK765 pKa = 10.77SDD767 pKa = 2.9GTAAGTDD774 pKa = 3.48IIGTDD779 pKa = 3.52GQAPNLGLGSFSLTAVDD796 pKa = 3.25NTLYY800 pKa = 10.55FVANDD805 pKa = 4.7PISGLEE811 pKa = 3.75LWKK814 pKa = 9.82TDD816 pKa = 3.05GTFANTAVVRR826 pKa = 11.84NISSGAADD834 pKa = 3.89SLPTGLVNFNGTLAFAASDD853 pKa = 4.42GINRR857 pKa = 11.84EE858 pKa = 3.93VWFSNGTEE866 pKa = 3.95LNTRR870 pKa = 11.84KK871 pKa = 9.98VSNINPTGNANPASLTVVGTKK892 pKa = 10.57LFFTANGANGTEE904 pKa = 4.5LYY906 pKa = 10.85VII908 pKa = 4.86
Molecular weight: 96.33 kDa
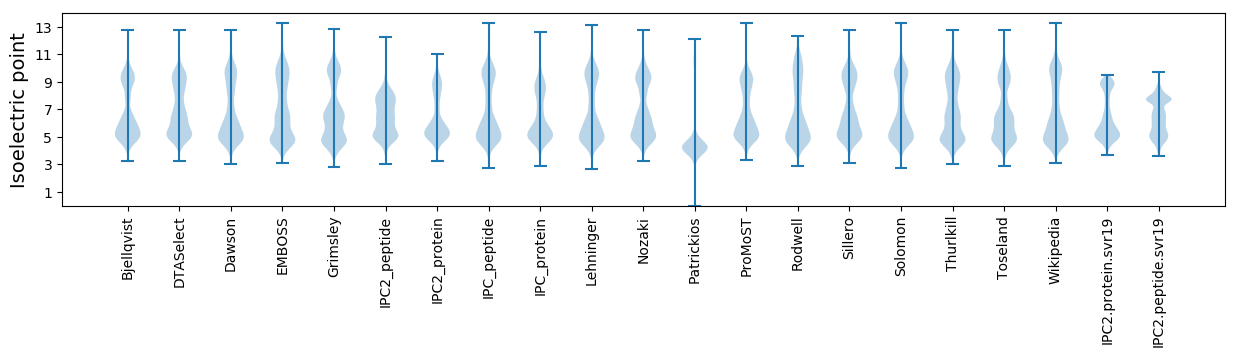
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8YS16|Q8YS16_NOSS1 Alr3276 protein OS=Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) OX=103690 GN=alr3276 PE=4 SV=1
MM1 pKa = 7.08QRR3 pKa = 11.84TLGGTNRR10 pKa = 11.84KK11 pKa = 9.1RR12 pKa = 11.84KK13 pKa = 7.65RR14 pKa = 11.84TSGFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TPDD26 pKa = 3.17GRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 8.83RR37 pKa = 11.84GRR39 pKa = 11.84HH40 pKa = 4.78RR41 pKa = 11.84LSVV44 pKa = 3.19
MM1 pKa = 7.08QRR3 pKa = 11.84TLGGTNRR10 pKa = 11.84KK11 pKa = 9.1RR12 pKa = 11.84KK13 pKa = 7.65RR14 pKa = 11.84TSGFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TPDD26 pKa = 3.17GRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 8.83RR37 pKa = 11.84GRR39 pKa = 11.84HH40 pKa = 4.78RR41 pKa = 11.84LSVV44 pKa = 3.19
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1955973 |
17 |
4936 |
322.2 |
35.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.998 ± 0.034 | 0.952 ± 0.013 |
4.81 ± 0.026 | 6.109 ± 0.034 |
3.948 ± 0.02 | 6.627 ± 0.043 |
1.832 ± 0.02 | 6.947 ± 0.026 |
4.786 ± 0.034 | 10.966 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.777 ± 0.016 | 4.635 ± 0.034 |
4.659 ± 0.024 | 5.555 ± 0.033 |
5.011 ± 0.031 | 6.43 ± 0.027 |
5.842 ± 0.037 | 6.619 ± 0.029 |
1.427 ± 0.012 | 3.069 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
