
Thaumarchaeota archaeon SCGC AB-539-E09
Taxonomy: cellular organisms; Archaea; TACK group; Thaumarchaeota; unclassified Thaumarchaeota
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
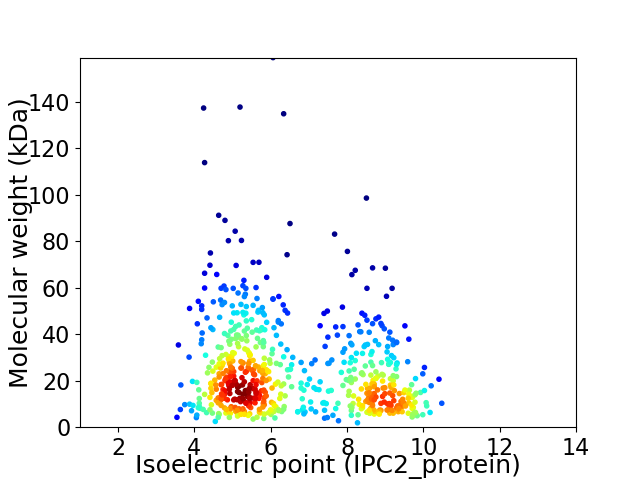
Virtual 2D-PAGE plot for 646 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M7TBZ5|M7TBZ5_9ARCH Nitrate/nitrite transporter OS=Thaumarchaeota archaeon SCGC AB-539-E09 OX=1198115 GN=MCGE09_00155 PE=4 SV=1
MM1 pKa = 7.27VLLPTCGSLFPDD13 pKa = 3.71EE14 pKa = 4.69EE15 pKa = 4.42MSLKK19 pKa = 10.13RR20 pKa = 11.84QYY22 pKa = 10.63IYY24 pKa = 11.3VVAICLSVIVAGAIIAYY41 pKa = 10.14NPFQEE46 pKa = 5.28KK47 pKa = 10.45EE48 pKa = 3.85DD49 pKa = 4.19DD50 pKa = 3.89NVFKK54 pKa = 10.55FPAGIPPSLGMEE66 pKa = 4.24YY67 pKa = 9.35ATTLGEE73 pKa = 4.11GDD75 pKa = 3.74VNSIQVSGQGSVSAKK90 pKa = 8.94ATQATLTLGAWTEE103 pKa = 4.18DD104 pKa = 3.41PVAAEE109 pKa = 3.9AVEE112 pKa = 4.91DD113 pKa = 3.84NALLMTSIIDD123 pKa = 4.24AIMGLGIDD131 pKa = 3.65EE132 pKa = 5.3DD133 pKa = 4.22KK134 pKa = 11.03IKK136 pKa = 10.05TVTYY140 pKa = 10.27SVSTNYY146 pKa = 10.31DD147 pKa = 2.77WEE149 pKa = 4.2TRR151 pKa = 11.84TIKK154 pKa = 10.55GYY156 pKa = 10.54RR157 pKa = 11.84VTNMIQVVVEE167 pKa = 4.77DD168 pKa = 3.51IDD170 pKa = 3.88MVGEE174 pKa = 4.48VIDD177 pKa = 3.56QAAAEE182 pKa = 4.46GANSIQGISFGISDD196 pKa = 4.5EE197 pKa = 4.24EE198 pKa = 4.41AKK200 pKa = 10.04TLANDD205 pKa = 4.17AYY207 pKa = 11.3VLALQNAQGKK217 pKa = 9.94ADD219 pKa = 5.66LIASTLDD226 pKa = 3.5LEE228 pKa = 4.65ITGVLYY234 pKa = 10.87VSEE237 pKa = 4.5SSYY240 pKa = 11.19TPYY243 pKa = 10.73SPYY246 pKa = 10.25RR247 pKa = 11.84SYY249 pKa = 11.7AEE251 pKa = 3.92EE252 pKa = 4.78AMDD255 pKa = 4.67SGFAAPTPIIEE266 pKa = 4.49GTLSVSVSVQVTFSFQQ282 pKa = 2.9
MM1 pKa = 7.27VLLPTCGSLFPDD13 pKa = 3.71EE14 pKa = 4.69EE15 pKa = 4.42MSLKK19 pKa = 10.13RR20 pKa = 11.84QYY22 pKa = 10.63IYY24 pKa = 11.3VVAICLSVIVAGAIIAYY41 pKa = 10.14NPFQEE46 pKa = 5.28KK47 pKa = 10.45EE48 pKa = 3.85DD49 pKa = 4.19DD50 pKa = 3.89NVFKK54 pKa = 10.55FPAGIPPSLGMEE66 pKa = 4.24YY67 pKa = 9.35ATTLGEE73 pKa = 4.11GDD75 pKa = 3.74VNSIQVSGQGSVSAKK90 pKa = 8.94ATQATLTLGAWTEE103 pKa = 4.18DD104 pKa = 3.41PVAAEE109 pKa = 3.9AVEE112 pKa = 4.91DD113 pKa = 3.84NALLMTSIIDD123 pKa = 4.24AIMGLGIDD131 pKa = 3.65EE132 pKa = 5.3DD133 pKa = 4.22KK134 pKa = 11.03IKK136 pKa = 10.05TVTYY140 pKa = 10.27SVSTNYY146 pKa = 10.31DD147 pKa = 2.77WEE149 pKa = 4.2TRR151 pKa = 11.84TIKK154 pKa = 10.55GYY156 pKa = 10.54RR157 pKa = 11.84VTNMIQVVVEE167 pKa = 4.77DD168 pKa = 3.51IDD170 pKa = 3.88MVGEE174 pKa = 4.48VIDD177 pKa = 3.56QAAAEE182 pKa = 4.46GANSIQGISFGISDD196 pKa = 4.5EE197 pKa = 4.24EE198 pKa = 4.41AKK200 pKa = 10.04TLANDD205 pKa = 4.17AYY207 pKa = 11.3VLALQNAQGKK217 pKa = 9.94ADD219 pKa = 5.66LIASTLDD226 pKa = 3.5LEE228 pKa = 4.65ITGVLYY234 pKa = 10.87VSEE237 pKa = 4.5SSYY240 pKa = 11.19TPYY243 pKa = 10.73SPYY246 pKa = 10.25RR247 pKa = 11.84SYY249 pKa = 11.7AEE251 pKa = 3.92EE252 pKa = 4.78AMDD255 pKa = 4.67SGFAAPTPIIEE266 pKa = 4.49GTLSVSVSVQVTFSFQQ282 pKa = 2.9
Molecular weight: 30.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M7TAJ0|M7TAJ0_9ARCH Extracellular solute-binding protein family 5 (Fragment) OS=Thaumarchaeota archaeon SCGC AB-539-E09 OX=1198115 GN=MCGE09_00563 PE=4 SV=1
MM1 pKa = 7.71RR2 pKa = 11.84LCSRR6 pKa = 11.84EE7 pKa = 3.74IGVGFSAFRR16 pKa = 11.84MLTGYY21 pKa = 11.15LNMPGRR27 pKa = 11.84TLGYY31 pKa = 10.01RR32 pKa = 11.84GIKK35 pKa = 9.74KK36 pKa = 9.45EE37 pKa = 3.77RR38 pKa = 11.84LYY40 pKa = 9.77MHH42 pKa = 7.19KK43 pKa = 10.51FFFNCC48 pKa = 4.15
MM1 pKa = 7.71RR2 pKa = 11.84LCSRR6 pKa = 11.84EE7 pKa = 3.74IGVGFSAFRR16 pKa = 11.84MLTGYY21 pKa = 11.15LNMPGRR27 pKa = 11.84TLGYY31 pKa = 10.01RR32 pKa = 11.84GIKK35 pKa = 9.74KK36 pKa = 9.45EE37 pKa = 3.77RR38 pKa = 11.84LYY40 pKa = 9.77MHH42 pKa = 7.19KK43 pKa = 10.51FFFNCC48 pKa = 4.15
Molecular weight: 5.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
143127 |
17 |
1365 |
221.6 |
25.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.682 ± 0.11 | 1.029 ± 0.034 |
5.522 ± 0.087 | 7.265 ± 0.131 |
4.258 ± 0.091 | 6.956 ± 0.102 |
1.856 ± 0.048 | 8.091 ± 0.12 |
6.354 ± 0.114 | 9.712 ± 0.105 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.692 ± 0.058 | 4.416 ± 0.097 |
4.193 ± 0.078 | 2.502 ± 0.056 |
5.252 ± 0.105 | 6.621 ± 0.085 |
5.238 ± 0.084 | 7.01 ± 0.102 |
1.348 ± 0.044 | 4.0 ± 0.08 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
