
Changjiang tombus-like virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.06
Get precalculated fractions of proteins
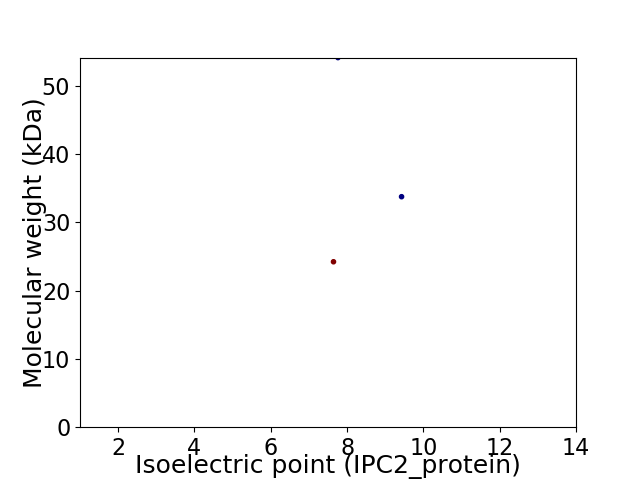
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFY4|A0A1L3KFY4_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 7 OX=1922821 PE=4 SV=1
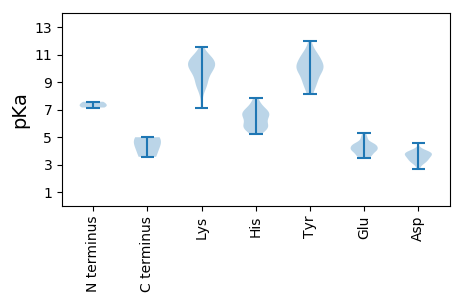
MM1 pKa = 7.14QWITANAFPCPPPPGLISSIFGEE24 pKa = 4.57VTITTYY30 pKa = 11.24GGTKK34 pKa = 9.91RR35 pKa = 11.84VWDD38 pKa = 3.45SRR40 pKa = 11.84KK41 pKa = 8.68IRR43 pKa = 11.84KK44 pKa = 7.09WAYY47 pKa = 9.15GAGAAASAALSLVALTAAGCGTSARR72 pKa = 11.84SLALRR77 pKa = 11.84TGAALLGVQAGAWLRR92 pKa = 11.84LHH94 pKa = 6.8YY95 pKa = 10.3DD96 pKa = 3.12AASTVPEE103 pKa = 4.01IDD105 pKa = 3.77EE106 pKa = 4.9LLAHH110 pKa = 6.62CADD113 pKa = 3.76IEE115 pKa = 4.32EE116 pKa = 4.44VTAVEE121 pKa = 4.49GKK123 pKa = 9.12PASEE127 pKa = 3.7AAEE130 pKa = 4.14ATKK133 pKa = 9.43TAANIVRR140 pKa = 11.84NWADD144 pKa = 2.81KK145 pKa = 10.78AYY147 pKa = 9.98AHH149 pKa = 6.85FGRR152 pKa = 11.84RR153 pKa = 11.84LEE155 pKa = 4.11DD156 pKa = 3.17TKK158 pKa = 11.55ADD160 pKa = 4.07RR161 pKa = 11.84DD162 pKa = 3.88ALGFWLARR170 pKa = 11.84QMKK173 pKa = 8.7EE174 pKa = 3.45GSVRR178 pKa = 11.84DD179 pKa = 3.67RR180 pKa = 11.84DD181 pKa = 3.45IAQYY185 pKa = 10.92LPLIVEE191 pKa = 4.36VCLLPSFAEE200 pKa = 4.03LTAQEE205 pKa = 4.16MRR207 pKa = 11.84RR208 pKa = 11.84SRR210 pKa = 11.84VANILRR216 pKa = 11.84RR217 pKa = 11.84EE218 pKa = 4.05CHH220 pKa = 5.72PPKK223 pKa = 10.76VV224 pKa = 3.56
MM1 pKa = 7.14QWITANAFPCPPPPGLISSIFGEE24 pKa = 4.57VTITTYY30 pKa = 11.24GGTKK34 pKa = 9.91RR35 pKa = 11.84VWDD38 pKa = 3.45SRR40 pKa = 11.84KK41 pKa = 8.68IRR43 pKa = 11.84KK44 pKa = 7.09WAYY47 pKa = 9.15GAGAAASAALSLVALTAAGCGTSARR72 pKa = 11.84SLALRR77 pKa = 11.84TGAALLGVQAGAWLRR92 pKa = 11.84LHH94 pKa = 6.8YY95 pKa = 10.3DD96 pKa = 3.12AASTVPEE103 pKa = 4.01IDD105 pKa = 3.77EE106 pKa = 4.9LLAHH110 pKa = 6.62CADD113 pKa = 3.76IEE115 pKa = 4.32EE116 pKa = 4.44VTAVEE121 pKa = 4.49GKK123 pKa = 9.12PASEE127 pKa = 3.7AAEE130 pKa = 4.14ATKK133 pKa = 9.43TAANIVRR140 pKa = 11.84NWADD144 pKa = 2.81KK145 pKa = 10.78AYY147 pKa = 9.98AHH149 pKa = 6.85FGRR152 pKa = 11.84RR153 pKa = 11.84LEE155 pKa = 4.11DD156 pKa = 3.17TKK158 pKa = 11.55ADD160 pKa = 4.07RR161 pKa = 11.84DD162 pKa = 3.88ALGFWLARR170 pKa = 11.84QMKK173 pKa = 8.7EE174 pKa = 3.45GSVRR178 pKa = 11.84DD179 pKa = 3.67RR180 pKa = 11.84DD181 pKa = 3.45IAQYY185 pKa = 10.92LPLIVEE191 pKa = 4.36VCLLPSFAEE200 pKa = 4.03LTAQEE205 pKa = 4.16MRR207 pKa = 11.84RR208 pKa = 11.84SRR210 pKa = 11.84VANILRR216 pKa = 11.84RR217 pKa = 11.84EE218 pKa = 4.05CHH220 pKa = 5.72PPKK223 pKa = 10.76VV224 pKa = 3.56
Molecular weight: 24.23 kDa
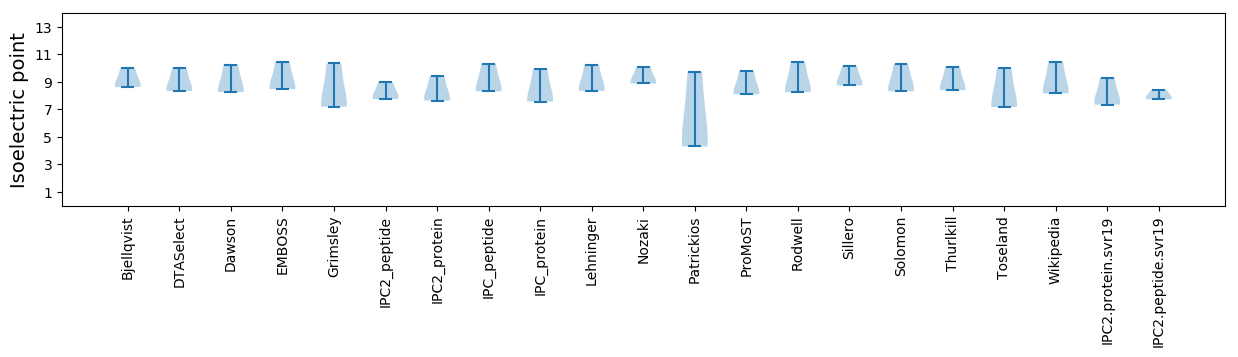
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG41|A0A1L3KG41_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 7 OX=1922821 PE=4 SV=1
MM1 pKa = 7.57ARR3 pKa = 11.84NKK5 pKa = 8.73ITNKK9 pKa = 10.35SPMTARR15 pKa = 11.84NPGGNNRR22 pKa = 11.84GPRR25 pKa = 11.84KK26 pKa = 9.6RR27 pKa = 11.84AGPKK31 pKa = 9.33PKK33 pKa = 9.9RR34 pKa = 11.84ANARR38 pKa = 11.84IGARR42 pKa = 11.84ALDD45 pKa = 3.89ASAAAYY51 pKa = 9.82ARR53 pKa = 11.84LLADD57 pKa = 3.36PCGAPLVHH65 pKa = 6.42PTYY68 pKa = 10.69SGSEE72 pKa = 3.62GGYY75 pKa = 10.23LIRR78 pKa = 11.84AEE80 pKa = 4.26SFVTPGVGATEE91 pKa = 3.9NAGYY95 pKa = 9.82FSWVPGLIGNAAGIQVGAAAGAGTSVAANNFGQNAPGFNFLRR137 pKa = 11.84DD138 pKa = 3.49TASGARR144 pKa = 11.84CVAACLKK151 pKa = 10.36VSYY154 pKa = 9.85PGTEE158 pKa = 3.55AGRR161 pKa = 11.84AGRR164 pKa = 11.84VHH166 pKa = 7.47FGQTSGGSIIQGVGYY181 pKa = 8.12TADD184 pKa = 3.47NVAVLLPHH192 pKa = 5.57YY193 pKa = 8.75TRR195 pKa = 11.84TPAQEE200 pKa = 4.86FEE202 pKa = 5.13LVWKK206 pKa = 10.16PNDD209 pKa = 3.46ADD211 pKa = 3.58QLFRR215 pKa = 11.84DD216 pKa = 4.58PSALTSEE223 pKa = 4.44EE224 pKa = 4.32SKK226 pKa = 10.67SARR229 pKa = 11.84AALTVAWAGLPVATGLTFRR248 pKa = 11.84LTAVYY253 pKa = 9.18EE254 pKa = 4.1WQPASADD261 pKa = 3.73GLSVPNMSRR270 pKa = 11.84SPSEE274 pKa = 3.74NTLDD278 pKa = 3.59NVINFLTRR286 pKa = 11.84QGFTFVRR293 pKa = 11.84GMAMAGTSGLISGISNHH310 pKa = 6.13FGIMSAGVNTRR321 pKa = 11.84RR322 pKa = 11.84LAMM325 pKa = 4.55
MM1 pKa = 7.57ARR3 pKa = 11.84NKK5 pKa = 8.73ITNKK9 pKa = 10.35SPMTARR15 pKa = 11.84NPGGNNRR22 pKa = 11.84GPRR25 pKa = 11.84KK26 pKa = 9.6RR27 pKa = 11.84AGPKK31 pKa = 9.33PKK33 pKa = 9.9RR34 pKa = 11.84ANARR38 pKa = 11.84IGARR42 pKa = 11.84ALDD45 pKa = 3.89ASAAAYY51 pKa = 9.82ARR53 pKa = 11.84LLADD57 pKa = 3.36PCGAPLVHH65 pKa = 6.42PTYY68 pKa = 10.69SGSEE72 pKa = 3.62GGYY75 pKa = 10.23LIRR78 pKa = 11.84AEE80 pKa = 4.26SFVTPGVGATEE91 pKa = 3.9NAGYY95 pKa = 9.82FSWVPGLIGNAAGIQVGAAAGAGTSVAANNFGQNAPGFNFLRR137 pKa = 11.84DD138 pKa = 3.49TASGARR144 pKa = 11.84CVAACLKK151 pKa = 10.36VSYY154 pKa = 9.85PGTEE158 pKa = 3.55AGRR161 pKa = 11.84AGRR164 pKa = 11.84VHH166 pKa = 7.47FGQTSGGSIIQGVGYY181 pKa = 8.12TADD184 pKa = 3.47NVAVLLPHH192 pKa = 5.57YY193 pKa = 8.75TRR195 pKa = 11.84TPAQEE200 pKa = 4.86FEE202 pKa = 5.13LVWKK206 pKa = 10.16PNDD209 pKa = 3.46ADD211 pKa = 3.58QLFRR215 pKa = 11.84DD216 pKa = 4.58PSALTSEE223 pKa = 4.44EE224 pKa = 4.32SKK226 pKa = 10.67SARR229 pKa = 11.84AALTVAWAGLPVATGLTFRR248 pKa = 11.84LTAVYY253 pKa = 9.18EE254 pKa = 4.1WQPASADD261 pKa = 3.73GLSVPNMSRR270 pKa = 11.84SPSEE274 pKa = 3.74NTLDD278 pKa = 3.59NVINFLTRR286 pKa = 11.84QGFTFVRR293 pKa = 11.84GMAMAGTSGLISGISNHH310 pKa = 6.13FGIMSAGVNTRR321 pKa = 11.84RR322 pKa = 11.84LAMM325 pKa = 4.55
Molecular weight: 33.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1035 |
224 |
486 |
345.0 |
37.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.754 ± 2.208 | 2.222 ± 0.552 |
3.865 ± 0.442 | 5.024 ± 0.793 |
4.155 ± 0.671 | 9.372 ± 1.177 |
2.415 ± 0.609 | 3.865 ± 0.342 |
3.478 ± 0.409 | 7.44 ± 0.616 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.609 ± 0.524 | 4.058 ± 0.96 |
5.7 ± 0.271 | 2.609 ± 0.159 |
7.633 ± 0.23 | 6.28 ± 0.469 |
5.411 ± 0.723 | 6.763 ± 0.477 |
1.643 ± 0.333 | 2.705 ± 0.217 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
