
Qingshengfaniella alkalisoli
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Qingshengfaniella
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
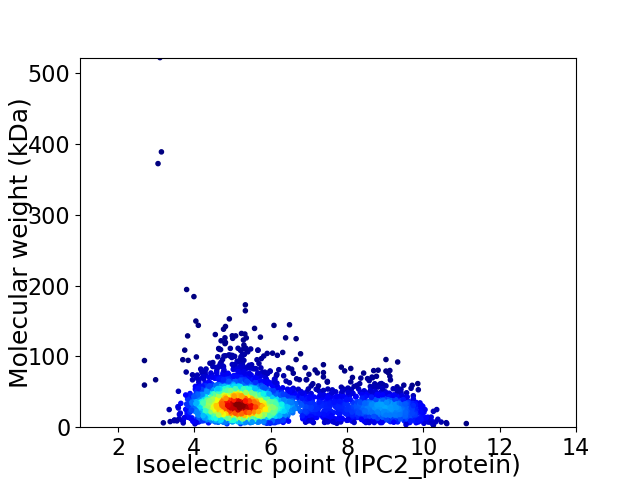
Virtual 2D-PAGE plot for 3573 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
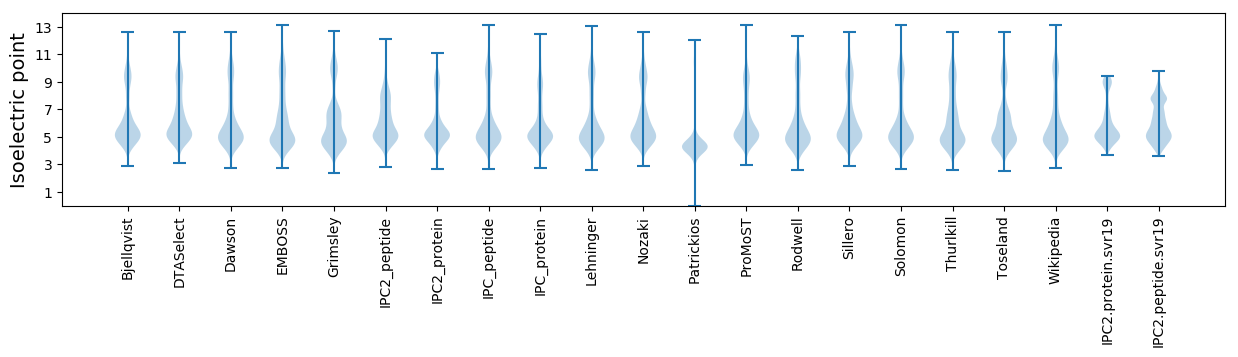
Protein with the lowest isoelectric point:
>tr|A0A5B8IUC8|A0A5B8IUC8_9RHOB Uncharacterized protein OS=Qingshengfaniella alkalisoli OX=2599296 GN=FPZ52_06130 PE=4 SV=1
MM1 pKa = 7.75RR2 pKa = 11.84LYY4 pKa = 10.84ASCAALALGLAAGGAHH20 pKa = 5.93AQSGEE25 pKa = 3.85ITVWSWNIAASSLEE39 pKa = 4.03SVIEE43 pKa = 4.22GFNAEE48 pKa = 4.26YY49 pKa = 10.26PDD51 pKa = 3.55VTVTVEE57 pKa = 4.39DD58 pKa = 4.5LGNQQVFDD66 pKa = 3.86RR67 pKa = 11.84TLAGCAAGGSGLPDD81 pKa = 3.07VLSIEE86 pKa = 4.02NHH88 pKa = 4.75EE89 pKa = 5.05AEE91 pKa = 4.61IFWAQFPDD99 pKa = 3.86CFANLRR105 pKa = 11.84DD106 pKa = 3.67LGYY109 pKa = 10.73DD110 pKa = 3.49EE111 pKa = 5.43EE112 pKa = 5.26SSARR116 pKa = 11.84FPDD119 pKa = 4.97FKK121 pKa = 10.47RR122 pKa = 11.84TEE124 pKa = 4.17LEE126 pKa = 4.2VGDD129 pKa = 4.1VAYY132 pKa = 10.7AMPWDD137 pKa = 4.19SGPVAMFYY145 pKa = 10.74RR146 pKa = 11.84RR147 pKa = 11.84DD148 pKa = 3.71FYY150 pKa = 11.72DD151 pKa = 3.32NAGVDD156 pKa = 3.64PASIQTWDD164 pKa = 3.76DD165 pKa = 4.49FIAAGKK171 pKa = 10.24QIMEE175 pKa = 4.59ANDD178 pKa = 3.88GVTMTNADD186 pKa = 3.96LNGDD190 pKa = 4.03TEE192 pKa = 4.61WFRR195 pKa = 11.84MIANEE200 pKa = 3.9QGCGYY205 pKa = 9.95FSNDD209 pKa = 3.15GEE211 pKa = 5.03SITVNQPSCVAALDD225 pKa = 4.09VVKK228 pKa = 10.96SLVDD232 pKa = 3.64EE233 pKa = 4.93GLMTAADD240 pKa = 4.04WNEE243 pKa = 3.93KK244 pKa = 8.91IQNNNAGAVASQMYY258 pKa = 8.99GGWYY262 pKa = 9.51EE263 pKa = 3.87GTIRR267 pKa = 11.84STAPEE272 pKa = 4.17DD273 pKa = 3.75QAGQWGVYY281 pKa = 10.01LMPSLSADD289 pKa = 3.46GAHH292 pKa = 6.93AANLGGSSLAIPANSEE308 pKa = 3.8NKK310 pKa = 8.19EE311 pKa = 4.01AAWAYY316 pKa = 10.69VNYY319 pKa = 10.87ALGTNDD325 pKa = 3.97GQVTMLKK332 pKa = 10.29SYY334 pKa = 11.1GLVPSLLSALEE345 pKa = 4.12DD346 pKa = 4.4PYY348 pKa = 11.78VQDD351 pKa = 5.64GLPFWGDD358 pKa = 3.23QPVWEE363 pKa = 5.17TILGTLDD370 pKa = 3.31QINPMRR376 pKa = 11.84GTSFFGDD383 pKa = 3.33ADD385 pKa = 3.84QIVQTVQTQYY395 pKa = 11.87LNGGYY400 pKa = 10.46DD401 pKa = 3.37SAQAALDD408 pKa = 3.91DD409 pKa = 3.97AAKK412 pKa = 10.37QIEE415 pKa = 5.23LVTGIPAAQQ424 pKa = 3.19
MM1 pKa = 7.75RR2 pKa = 11.84LYY4 pKa = 10.84ASCAALALGLAAGGAHH20 pKa = 5.93AQSGEE25 pKa = 3.85ITVWSWNIAASSLEE39 pKa = 4.03SVIEE43 pKa = 4.22GFNAEE48 pKa = 4.26YY49 pKa = 10.26PDD51 pKa = 3.55VTVTVEE57 pKa = 4.39DD58 pKa = 4.5LGNQQVFDD66 pKa = 3.86RR67 pKa = 11.84TLAGCAAGGSGLPDD81 pKa = 3.07VLSIEE86 pKa = 4.02NHH88 pKa = 4.75EE89 pKa = 5.05AEE91 pKa = 4.61IFWAQFPDD99 pKa = 3.86CFANLRR105 pKa = 11.84DD106 pKa = 3.67LGYY109 pKa = 10.73DD110 pKa = 3.49EE111 pKa = 5.43EE112 pKa = 5.26SSARR116 pKa = 11.84FPDD119 pKa = 4.97FKK121 pKa = 10.47RR122 pKa = 11.84TEE124 pKa = 4.17LEE126 pKa = 4.2VGDD129 pKa = 4.1VAYY132 pKa = 10.7AMPWDD137 pKa = 4.19SGPVAMFYY145 pKa = 10.74RR146 pKa = 11.84RR147 pKa = 11.84DD148 pKa = 3.71FYY150 pKa = 11.72DD151 pKa = 3.32NAGVDD156 pKa = 3.64PASIQTWDD164 pKa = 3.76DD165 pKa = 4.49FIAAGKK171 pKa = 10.24QIMEE175 pKa = 4.59ANDD178 pKa = 3.88GVTMTNADD186 pKa = 3.96LNGDD190 pKa = 4.03TEE192 pKa = 4.61WFRR195 pKa = 11.84MIANEE200 pKa = 3.9QGCGYY205 pKa = 9.95FSNDD209 pKa = 3.15GEE211 pKa = 5.03SITVNQPSCVAALDD225 pKa = 4.09VVKK228 pKa = 10.96SLVDD232 pKa = 3.64EE233 pKa = 4.93GLMTAADD240 pKa = 4.04WNEE243 pKa = 3.93KK244 pKa = 8.91IQNNNAGAVASQMYY258 pKa = 8.99GGWYY262 pKa = 9.51EE263 pKa = 3.87GTIRR267 pKa = 11.84STAPEE272 pKa = 4.17DD273 pKa = 3.75QAGQWGVYY281 pKa = 10.01LMPSLSADD289 pKa = 3.46GAHH292 pKa = 6.93AANLGGSSLAIPANSEE308 pKa = 3.8NKK310 pKa = 8.19EE311 pKa = 4.01AAWAYY316 pKa = 10.69VNYY319 pKa = 10.87ALGTNDD325 pKa = 3.97GQVTMLKK332 pKa = 10.29SYY334 pKa = 11.1GLVPSLLSALEE345 pKa = 4.12DD346 pKa = 4.4PYY348 pKa = 11.78VQDD351 pKa = 5.64GLPFWGDD358 pKa = 3.23QPVWEE363 pKa = 5.17TILGTLDD370 pKa = 3.31QINPMRR376 pKa = 11.84GTSFFGDD383 pKa = 3.33ADD385 pKa = 3.84QIVQTVQTQYY395 pKa = 11.87LNGGYY400 pKa = 10.46DD401 pKa = 3.37SAQAALDD408 pKa = 3.91DD409 pKa = 3.97AAKK412 pKa = 10.37QIEE415 pKa = 5.23LVTGIPAAQQ424 pKa = 3.19
Molecular weight: 45.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B8J6R1|A0A5B8J6R1_9RHOB DUF882 domain-containing protein OS=Qingshengfaniella alkalisoli OX=2599296 GN=FPZ52_11175 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1157484 |
25 |
5139 |
324.0 |
35.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.322 ± 0.051 | 0.883 ± 0.014 |
6.432 ± 0.057 | 5.911 ± 0.036 |
3.761 ± 0.026 | 8.452 ± 0.048 |
2.016 ± 0.021 | 5.482 ± 0.029 |
3.181 ± 0.038 | 10.013 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.698 ± 0.021 | 2.8 ± 0.024 |
4.967 ± 0.025 | 3.291 ± 0.022 |
6.574 ± 0.056 | 5.612 ± 0.03 |
5.593 ± 0.056 | 7.38 ± 0.039 |
1.357 ± 0.019 | 2.276 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
