
Carp edema virus
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Pokkesviricetes; Chitovirales; Poxviridae; unclassified Poxviridae
Average proteome isoelectric point is 5.4
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A5B7QBP8|A0A5B7QBP8_9POXV Major core protein 4a precursor OS=Carp edema virus OX=1653087 GN=P4a PE=3 SV=1
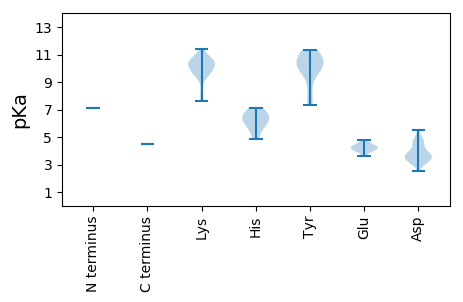
MM1 pKa = 7.1VATAAARR8 pKa = 11.84DD9 pKa = 3.76PEE11 pKa = 4.63LRR13 pKa = 11.84DD14 pKa = 3.37SNYY17 pKa = 10.35VMAAPAANPFANMVKK32 pKa = 10.26FDD34 pKa = 4.47IPDD37 pKa = 3.7IYY39 pKa = 11.05QFAEE43 pKa = 3.93IGLQQIKK50 pKa = 7.88MTTLHH55 pKa = 6.06YY56 pKa = 10.66CDD58 pKa = 4.84FVSKK62 pKa = 10.46VLPHH66 pKa = 6.55PNYY69 pKa = 9.99VVKK72 pKa = 10.72KK73 pKa = 10.12DD74 pKa = 3.12IGYY77 pKa = 10.17VDD79 pKa = 4.9TIPEE83 pKa = 3.92QVKK86 pKa = 9.9IANAMFWKK94 pKa = 10.43GIQIDD99 pKa = 4.22DD100 pKa = 3.85WNKK103 pKa = 9.57MNVFDD108 pKa = 5.49KK109 pKa = 11.3TMTSNLALVLGLYY122 pKa = 10.72DD123 pKa = 3.25EE124 pKa = 4.61TVYY127 pKa = 10.83FKK129 pKa = 11.41NSTNKK134 pKa = 9.92FEE136 pKa = 4.3ITFCGILDD144 pKa = 3.95QIPQGVAVKK153 pKa = 9.74RR154 pKa = 11.84VCFLPYY160 pKa = 10.48KK161 pKa = 10.2LGNATIYY168 pKa = 8.5KK169 pKa = 7.62TLKK172 pKa = 10.34HH173 pKa = 6.25VSTQIWEE180 pKa = 4.14GFYY183 pKa = 11.28ANGNLITNIKK193 pKa = 9.57QLQKK197 pKa = 9.68VTNDD201 pKa = 3.13EE202 pKa = 4.34KK203 pKa = 10.87KK204 pKa = 9.05TLGLTKK210 pKa = 9.47TQYY213 pKa = 11.28TNFIDD218 pKa = 4.77NINLSTLDD226 pKa = 3.53TPYY229 pKa = 11.31KK230 pKa = 10.58LLAFYY235 pKa = 7.49CTCRR239 pKa = 11.84YY240 pKa = 9.72KK241 pKa = 11.4DD242 pKa = 3.83NDD244 pKa = 3.6PAANKK249 pKa = 9.59VLYY252 pKa = 9.9KK253 pKa = 10.32IPSNKK258 pKa = 9.64AVLNLIWILLTKK270 pKa = 10.16LGYY273 pKa = 10.2VIRR276 pKa = 11.84STTNTKK282 pKa = 9.32TDD284 pKa = 3.29EE285 pKa = 4.26TSFKK289 pKa = 10.68VFPSFTKK296 pKa = 9.66QSLRR300 pKa = 11.84AKK302 pKa = 10.31VIAALRR308 pKa = 11.84KK309 pKa = 10.01DD310 pKa = 3.78GNNVEE315 pKa = 4.76AADD318 pKa = 3.85TLINNCEE325 pKa = 3.87PLILFIYY332 pKa = 10.37NFLSYY337 pKa = 10.54PSATKK342 pKa = 10.39ACADD346 pKa = 3.11IAGAGKK352 pKa = 10.39GSKK355 pKa = 10.03EE356 pKa = 4.04DD357 pKa = 3.56TSGGDD362 pKa = 3.39TLSVEE367 pKa = 4.08NDD369 pKa = 4.42PIPFYY374 pKa = 11.25HH375 pKa = 7.11NINIQEE381 pKa = 4.33LLLTEE386 pKa = 4.47SGEE389 pKa = 4.05EE390 pKa = 4.04LKK392 pKa = 10.9NCRR395 pKa = 11.84PFKK398 pKa = 10.68NCIDD402 pKa = 3.76NFKK405 pKa = 10.19KK406 pKa = 10.39AVYY409 pKa = 7.31EE410 pKa = 4.08APMKK414 pKa = 10.87DD415 pKa = 4.48DD416 pKa = 3.38ILEE419 pKa = 4.24FADD422 pKa = 4.66SIKK425 pKa = 10.76SSYY428 pKa = 9.76PEE430 pKa = 3.61HH431 pKa = 6.72LRR433 pKa = 11.84WNLAVSGSVDD443 pKa = 3.27GSIINADD450 pKa = 3.83NIVDD454 pKa = 3.58IVEE457 pKa = 4.68KK458 pKa = 10.68SVQHH462 pKa = 6.47DD463 pKa = 3.55FNNIFRR469 pKa = 11.84SEE471 pKa = 4.33SEE473 pKa = 4.33DD474 pKa = 3.28VAAGYY479 pKa = 9.56GIKK482 pKa = 10.37DD483 pKa = 2.97IFEE486 pKa = 4.15QLEE489 pKa = 4.36SEE491 pKa = 4.39TTEE494 pKa = 4.32LEE496 pKa = 3.82SNYY499 pKa = 9.94IVDD502 pKa = 4.23EE503 pKa = 4.14LAKK506 pKa = 10.37QLVSSRR512 pKa = 11.84QIQRR516 pKa = 11.84MTTRR520 pKa = 11.84PGHH523 pKa = 4.86FLKK526 pKa = 10.8RR527 pKa = 11.84IPEE530 pKa = 3.81YY531 pKa = 10.75ALRR534 pKa = 11.84HH535 pKa = 5.43RR536 pKa = 11.84SMLKK540 pKa = 8.45TGEE543 pKa = 4.03DD544 pKa = 2.51VGYY547 pKa = 10.33IMNSMVSNMKK557 pKa = 10.35ALEE560 pKa = 4.13KK561 pKa = 9.67THH563 pKa = 6.09GVEE566 pKa = 3.73INADD570 pKa = 3.55VKK572 pKa = 10.8EE573 pKa = 4.55VIAKK577 pKa = 9.38QGNAIIKK584 pKa = 9.78NLYY587 pKa = 9.76SLYY590 pKa = 8.56KK591 pKa = 8.15TCVVVDD597 pKa = 5.01FTRR600 pKa = 11.84GLLEE604 pKa = 4.31FIGRR608 pKa = 11.84GGSANKK614 pKa = 10.01HH615 pKa = 5.74GLDD618 pKa = 3.36EE619 pKa = 4.29TDD621 pKa = 3.84IDD623 pKa = 4.65SEE625 pKa = 4.55TLNVIYY631 pKa = 10.73NHH633 pKa = 6.81IKK635 pKa = 10.31PSLTSNLNAYY645 pKa = 7.51QTTINKK651 pKa = 9.63IIEE654 pKa = 4.37KK655 pKa = 10.37VSGEE659 pKa = 4.17LGVQSEE665 pKa = 4.55EE666 pKa = 4.08LEE668 pKa = 4.25AANEE672 pKa = 4.04EE673 pKa = 4.52LFF675 pKa = 4.46
MM1 pKa = 7.1VATAAARR8 pKa = 11.84DD9 pKa = 3.76PEE11 pKa = 4.63LRR13 pKa = 11.84DD14 pKa = 3.37SNYY17 pKa = 10.35VMAAPAANPFANMVKK32 pKa = 10.26FDD34 pKa = 4.47IPDD37 pKa = 3.7IYY39 pKa = 11.05QFAEE43 pKa = 3.93IGLQQIKK50 pKa = 7.88MTTLHH55 pKa = 6.06YY56 pKa = 10.66CDD58 pKa = 4.84FVSKK62 pKa = 10.46VLPHH66 pKa = 6.55PNYY69 pKa = 9.99VVKK72 pKa = 10.72KK73 pKa = 10.12DD74 pKa = 3.12IGYY77 pKa = 10.17VDD79 pKa = 4.9TIPEE83 pKa = 3.92QVKK86 pKa = 9.9IANAMFWKK94 pKa = 10.43GIQIDD99 pKa = 4.22DD100 pKa = 3.85WNKK103 pKa = 9.57MNVFDD108 pKa = 5.49KK109 pKa = 11.3TMTSNLALVLGLYY122 pKa = 10.72DD123 pKa = 3.25EE124 pKa = 4.61TVYY127 pKa = 10.83FKK129 pKa = 11.41NSTNKK134 pKa = 9.92FEE136 pKa = 4.3ITFCGILDD144 pKa = 3.95QIPQGVAVKK153 pKa = 9.74RR154 pKa = 11.84VCFLPYY160 pKa = 10.48KK161 pKa = 10.2LGNATIYY168 pKa = 8.5KK169 pKa = 7.62TLKK172 pKa = 10.34HH173 pKa = 6.25VSTQIWEE180 pKa = 4.14GFYY183 pKa = 11.28ANGNLITNIKK193 pKa = 9.57QLQKK197 pKa = 9.68VTNDD201 pKa = 3.13EE202 pKa = 4.34KK203 pKa = 10.87KK204 pKa = 9.05TLGLTKK210 pKa = 9.47TQYY213 pKa = 11.28TNFIDD218 pKa = 4.77NINLSTLDD226 pKa = 3.53TPYY229 pKa = 11.31KK230 pKa = 10.58LLAFYY235 pKa = 7.49CTCRR239 pKa = 11.84YY240 pKa = 9.72KK241 pKa = 11.4DD242 pKa = 3.83NDD244 pKa = 3.6PAANKK249 pKa = 9.59VLYY252 pKa = 9.9KK253 pKa = 10.32IPSNKK258 pKa = 9.64AVLNLIWILLTKK270 pKa = 10.16LGYY273 pKa = 10.2VIRR276 pKa = 11.84STTNTKK282 pKa = 9.32TDD284 pKa = 3.29EE285 pKa = 4.26TSFKK289 pKa = 10.68VFPSFTKK296 pKa = 9.66QSLRR300 pKa = 11.84AKK302 pKa = 10.31VIAALRR308 pKa = 11.84KK309 pKa = 10.01DD310 pKa = 3.78GNNVEE315 pKa = 4.76AADD318 pKa = 3.85TLINNCEE325 pKa = 3.87PLILFIYY332 pKa = 10.37NFLSYY337 pKa = 10.54PSATKK342 pKa = 10.39ACADD346 pKa = 3.11IAGAGKK352 pKa = 10.39GSKK355 pKa = 10.03EE356 pKa = 4.04DD357 pKa = 3.56TSGGDD362 pKa = 3.39TLSVEE367 pKa = 4.08NDD369 pKa = 4.42PIPFYY374 pKa = 11.25HH375 pKa = 7.11NINIQEE381 pKa = 4.33LLLTEE386 pKa = 4.47SGEE389 pKa = 4.05EE390 pKa = 4.04LKK392 pKa = 10.9NCRR395 pKa = 11.84PFKK398 pKa = 10.68NCIDD402 pKa = 3.76NFKK405 pKa = 10.19KK406 pKa = 10.39AVYY409 pKa = 7.31EE410 pKa = 4.08APMKK414 pKa = 10.87DD415 pKa = 4.48DD416 pKa = 3.38ILEE419 pKa = 4.24FADD422 pKa = 4.66SIKK425 pKa = 10.76SSYY428 pKa = 9.76PEE430 pKa = 3.61HH431 pKa = 6.72LRR433 pKa = 11.84WNLAVSGSVDD443 pKa = 3.27GSIINADD450 pKa = 3.83NIVDD454 pKa = 3.58IVEE457 pKa = 4.68KK458 pKa = 10.68SVQHH462 pKa = 6.47DD463 pKa = 3.55FNNIFRR469 pKa = 11.84SEE471 pKa = 4.33SEE473 pKa = 4.33DD474 pKa = 3.28VAAGYY479 pKa = 9.56GIKK482 pKa = 10.37DD483 pKa = 2.97IFEE486 pKa = 4.15QLEE489 pKa = 4.36SEE491 pKa = 4.39TTEE494 pKa = 4.32LEE496 pKa = 3.82SNYY499 pKa = 9.94IVDD502 pKa = 4.23EE503 pKa = 4.14LAKK506 pKa = 10.37QLVSSRR512 pKa = 11.84QIQRR516 pKa = 11.84MTTRR520 pKa = 11.84PGHH523 pKa = 4.86FLKK526 pKa = 10.8RR527 pKa = 11.84IPEE530 pKa = 3.81YY531 pKa = 10.75ALRR534 pKa = 11.84HH535 pKa = 5.43RR536 pKa = 11.84SMLKK540 pKa = 8.45TGEE543 pKa = 4.03DD544 pKa = 2.51VGYY547 pKa = 10.33IMNSMVSNMKK557 pKa = 10.35ALEE560 pKa = 4.13KK561 pKa = 9.67THH563 pKa = 6.09GVEE566 pKa = 3.73INADD570 pKa = 3.55VKK572 pKa = 10.8EE573 pKa = 4.55VIAKK577 pKa = 9.38QGNAIIKK584 pKa = 9.78NLYY587 pKa = 9.76SLYY590 pKa = 8.56KK591 pKa = 8.15TCVVVDD597 pKa = 5.01FTRR600 pKa = 11.84GLLEE604 pKa = 4.31FIGRR608 pKa = 11.84GGSANKK614 pKa = 10.01HH615 pKa = 5.74GLDD618 pKa = 3.36EE619 pKa = 4.29TDD621 pKa = 3.84IDD623 pKa = 4.65SEE625 pKa = 4.55TLNVIYY631 pKa = 10.73NHH633 pKa = 6.81IKK635 pKa = 10.31PSLTSNLNAYY645 pKa = 7.51QTTINKK651 pKa = 9.63IIEE654 pKa = 4.37KK655 pKa = 10.37VSGEE659 pKa = 4.17LGVQSEE665 pKa = 4.55EE666 pKa = 4.08LEE668 pKa = 4.25AANEE672 pKa = 4.04EE673 pKa = 4.52LFF675 pKa = 4.46
Molecular weight: 75.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B7QBP8|A0A5B7QBP8_9POXV Major core protein 4a precursor OS=Carp edema virus OX=1653087 GN=P4a PE=3 SV=1
MM1 pKa = 7.1VATAAARR8 pKa = 11.84DD9 pKa = 3.76PEE11 pKa = 4.63LRR13 pKa = 11.84DD14 pKa = 3.37SNYY17 pKa = 10.35VMAAPAANPFANMVKK32 pKa = 10.26FDD34 pKa = 4.47IPDD37 pKa = 3.7IYY39 pKa = 11.05QFAEE43 pKa = 3.93IGLQQIKK50 pKa = 7.88MTTLHH55 pKa = 6.06YY56 pKa = 10.66CDD58 pKa = 4.84FVSKK62 pKa = 10.46VLPHH66 pKa = 6.55PNYY69 pKa = 9.99VVKK72 pKa = 10.72KK73 pKa = 10.12DD74 pKa = 3.12IGYY77 pKa = 10.17VDD79 pKa = 4.9TIPEE83 pKa = 3.92QVKK86 pKa = 9.9IANAMFWKK94 pKa = 10.43GIQIDD99 pKa = 4.22DD100 pKa = 3.85WNKK103 pKa = 9.57MNVFDD108 pKa = 5.49KK109 pKa = 11.3TMTSNLALVLGLYY122 pKa = 10.72DD123 pKa = 3.25EE124 pKa = 4.61TVYY127 pKa = 10.83FKK129 pKa = 11.41NSTNKK134 pKa = 9.92FEE136 pKa = 4.3ITFCGILDD144 pKa = 3.95QIPQGVAVKK153 pKa = 9.74RR154 pKa = 11.84VCFLPYY160 pKa = 10.48KK161 pKa = 10.2LGNATIYY168 pKa = 8.5KK169 pKa = 7.62TLKK172 pKa = 10.34HH173 pKa = 6.25VSTQIWEE180 pKa = 4.14GFYY183 pKa = 11.28ANGNLITNIKK193 pKa = 9.57QLQKK197 pKa = 9.68VTNDD201 pKa = 3.13EE202 pKa = 4.34KK203 pKa = 10.87KK204 pKa = 9.05TLGLTKK210 pKa = 9.47TQYY213 pKa = 11.28TNFIDD218 pKa = 4.77NINLSTLDD226 pKa = 3.53TPYY229 pKa = 11.31KK230 pKa = 10.58LLAFYY235 pKa = 7.49CTCRR239 pKa = 11.84YY240 pKa = 9.72KK241 pKa = 11.4DD242 pKa = 3.83NDD244 pKa = 3.6PAANKK249 pKa = 9.59VLYY252 pKa = 9.9KK253 pKa = 10.32IPSNKK258 pKa = 9.64AVLNLIWILLTKK270 pKa = 10.16LGYY273 pKa = 10.2VIRR276 pKa = 11.84STTNTKK282 pKa = 9.32TDD284 pKa = 3.29EE285 pKa = 4.26TSFKK289 pKa = 10.68VFPSFTKK296 pKa = 9.66QSLRR300 pKa = 11.84AKK302 pKa = 10.31VIAALRR308 pKa = 11.84KK309 pKa = 10.01DD310 pKa = 3.78GNNVEE315 pKa = 4.76AADD318 pKa = 3.85TLINNCEE325 pKa = 3.87PLILFIYY332 pKa = 10.37NFLSYY337 pKa = 10.54PSATKK342 pKa = 10.39ACADD346 pKa = 3.11IAGAGKK352 pKa = 10.39GSKK355 pKa = 10.03EE356 pKa = 4.04DD357 pKa = 3.56TSGGDD362 pKa = 3.39TLSVEE367 pKa = 4.08NDD369 pKa = 4.42PIPFYY374 pKa = 11.25HH375 pKa = 7.11NINIQEE381 pKa = 4.33LLLTEE386 pKa = 4.47SGEE389 pKa = 4.05EE390 pKa = 4.04LKK392 pKa = 10.9NCRR395 pKa = 11.84PFKK398 pKa = 10.68NCIDD402 pKa = 3.76NFKK405 pKa = 10.19KK406 pKa = 10.39AVYY409 pKa = 7.31EE410 pKa = 4.08APMKK414 pKa = 10.87DD415 pKa = 4.48DD416 pKa = 3.38ILEE419 pKa = 4.24FADD422 pKa = 4.66SIKK425 pKa = 10.76SSYY428 pKa = 9.76PEE430 pKa = 3.61HH431 pKa = 6.72LRR433 pKa = 11.84WNLAVSGSVDD443 pKa = 3.27GSIINADD450 pKa = 3.83NIVDD454 pKa = 3.58IVEE457 pKa = 4.68KK458 pKa = 10.68SVQHH462 pKa = 6.47DD463 pKa = 3.55FNNIFRR469 pKa = 11.84SEE471 pKa = 4.33SEE473 pKa = 4.33DD474 pKa = 3.28VAAGYY479 pKa = 9.56GIKK482 pKa = 10.37DD483 pKa = 2.97IFEE486 pKa = 4.15QLEE489 pKa = 4.36SEE491 pKa = 4.39TTEE494 pKa = 4.32LEE496 pKa = 3.82SNYY499 pKa = 9.94IVDD502 pKa = 4.23EE503 pKa = 4.14LAKK506 pKa = 10.37QLVSSRR512 pKa = 11.84QIQRR516 pKa = 11.84MTTRR520 pKa = 11.84PGHH523 pKa = 4.86FLKK526 pKa = 10.8RR527 pKa = 11.84IPEE530 pKa = 3.81YY531 pKa = 10.75ALRR534 pKa = 11.84HH535 pKa = 5.43RR536 pKa = 11.84SMLKK540 pKa = 8.45TGEE543 pKa = 4.03DD544 pKa = 2.51VGYY547 pKa = 10.33IMNSMVSNMKK557 pKa = 10.35ALEE560 pKa = 4.13KK561 pKa = 9.67THH563 pKa = 6.09GVEE566 pKa = 3.73INADD570 pKa = 3.55VKK572 pKa = 10.8EE573 pKa = 4.55VIAKK577 pKa = 9.38QGNAIIKK584 pKa = 9.78NLYY587 pKa = 9.76SLYY590 pKa = 8.56KK591 pKa = 8.15TCVVVDD597 pKa = 5.01FTRR600 pKa = 11.84GLLEE604 pKa = 4.31FIGRR608 pKa = 11.84GGSANKK614 pKa = 10.01HH615 pKa = 5.74GLDD618 pKa = 3.36EE619 pKa = 4.29TDD621 pKa = 3.84IDD623 pKa = 4.65SEE625 pKa = 4.55TLNVIYY631 pKa = 10.73NHH633 pKa = 6.81IKK635 pKa = 10.31PSLTSNLNAYY645 pKa = 7.51QTTINKK651 pKa = 9.63IIEE654 pKa = 4.37KK655 pKa = 10.37VSGEE659 pKa = 4.17LGVQSEE665 pKa = 4.55EE666 pKa = 4.08LEE668 pKa = 4.25AANEE672 pKa = 4.04EE673 pKa = 4.52LFF675 pKa = 4.46
MM1 pKa = 7.1VATAAARR8 pKa = 11.84DD9 pKa = 3.76PEE11 pKa = 4.63LRR13 pKa = 11.84DD14 pKa = 3.37SNYY17 pKa = 10.35VMAAPAANPFANMVKK32 pKa = 10.26FDD34 pKa = 4.47IPDD37 pKa = 3.7IYY39 pKa = 11.05QFAEE43 pKa = 3.93IGLQQIKK50 pKa = 7.88MTTLHH55 pKa = 6.06YY56 pKa = 10.66CDD58 pKa = 4.84FVSKK62 pKa = 10.46VLPHH66 pKa = 6.55PNYY69 pKa = 9.99VVKK72 pKa = 10.72KK73 pKa = 10.12DD74 pKa = 3.12IGYY77 pKa = 10.17VDD79 pKa = 4.9TIPEE83 pKa = 3.92QVKK86 pKa = 9.9IANAMFWKK94 pKa = 10.43GIQIDD99 pKa = 4.22DD100 pKa = 3.85WNKK103 pKa = 9.57MNVFDD108 pKa = 5.49KK109 pKa = 11.3TMTSNLALVLGLYY122 pKa = 10.72DD123 pKa = 3.25EE124 pKa = 4.61TVYY127 pKa = 10.83FKK129 pKa = 11.41NSTNKK134 pKa = 9.92FEE136 pKa = 4.3ITFCGILDD144 pKa = 3.95QIPQGVAVKK153 pKa = 9.74RR154 pKa = 11.84VCFLPYY160 pKa = 10.48KK161 pKa = 10.2LGNATIYY168 pKa = 8.5KK169 pKa = 7.62TLKK172 pKa = 10.34HH173 pKa = 6.25VSTQIWEE180 pKa = 4.14GFYY183 pKa = 11.28ANGNLITNIKK193 pKa = 9.57QLQKK197 pKa = 9.68VTNDD201 pKa = 3.13EE202 pKa = 4.34KK203 pKa = 10.87KK204 pKa = 9.05TLGLTKK210 pKa = 9.47TQYY213 pKa = 11.28TNFIDD218 pKa = 4.77NINLSTLDD226 pKa = 3.53TPYY229 pKa = 11.31KK230 pKa = 10.58LLAFYY235 pKa = 7.49CTCRR239 pKa = 11.84YY240 pKa = 9.72KK241 pKa = 11.4DD242 pKa = 3.83NDD244 pKa = 3.6PAANKK249 pKa = 9.59VLYY252 pKa = 9.9KK253 pKa = 10.32IPSNKK258 pKa = 9.64AVLNLIWILLTKK270 pKa = 10.16LGYY273 pKa = 10.2VIRR276 pKa = 11.84STTNTKK282 pKa = 9.32TDD284 pKa = 3.29EE285 pKa = 4.26TSFKK289 pKa = 10.68VFPSFTKK296 pKa = 9.66QSLRR300 pKa = 11.84AKK302 pKa = 10.31VIAALRR308 pKa = 11.84KK309 pKa = 10.01DD310 pKa = 3.78GNNVEE315 pKa = 4.76AADD318 pKa = 3.85TLINNCEE325 pKa = 3.87PLILFIYY332 pKa = 10.37NFLSYY337 pKa = 10.54PSATKK342 pKa = 10.39ACADD346 pKa = 3.11IAGAGKK352 pKa = 10.39GSKK355 pKa = 10.03EE356 pKa = 4.04DD357 pKa = 3.56TSGGDD362 pKa = 3.39TLSVEE367 pKa = 4.08NDD369 pKa = 4.42PIPFYY374 pKa = 11.25HH375 pKa = 7.11NINIQEE381 pKa = 4.33LLLTEE386 pKa = 4.47SGEE389 pKa = 4.05EE390 pKa = 4.04LKK392 pKa = 10.9NCRR395 pKa = 11.84PFKK398 pKa = 10.68NCIDD402 pKa = 3.76NFKK405 pKa = 10.19KK406 pKa = 10.39AVYY409 pKa = 7.31EE410 pKa = 4.08APMKK414 pKa = 10.87DD415 pKa = 4.48DD416 pKa = 3.38ILEE419 pKa = 4.24FADD422 pKa = 4.66SIKK425 pKa = 10.76SSYY428 pKa = 9.76PEE430 pKa = 3.61HH431 pKa = 6.72LRR433 pKa = 11.84WNLAVSGSVDD443 pKa = 3.27GSIINADD450 pKa = 3.83NIVDD454 pKa = 3.58IVEE457 pKa = 4.68KK458 pKa = 10.68SVQHH462 pKa = 6.47DD463 pKa = 3.55FNNIFRR469 pKa = 11.84SEE471 pKa = 4.33SEE473 pKa = 4.33DD474 pKa = 3.28VAAGYY479 pKa = 9.56GIKK482 pKa = 10.37DD483 pKa = 2.97IFEE486 pKa = 4.15QLEE489 pKa = 4.36SEE491 pKa = 4.39TTEE494 pKa = 4.32LEE496 pKa = 3.82SNYY499 pKa = 9.94IVDD502 pKa = 4.23EE503 pKa = 4.14LAKK506 pKa = 10.37QLVSSRR512 pKa = 11.84QIQRR516 pKa = 11.84MTTRR520 pKa = 11.84PGHH523 pKa = 4.86FLKK526 pKa = 10.8RR527 pKa = 11.84IPEE530 pKa = 3.81YY531 pKa = 10.75ALRR534 pKa = 11.84HH535 pKa = 5.43RR536 pKa = 11.84SMLKK540 pKa = 8.45TGEE543 pKa = 4.03DD544 pKa = 2.51VGYY547 pKa = 10.33IMNSMVSNMKK557 pKa = 10.35ALEE560 pKa = 4.13KK561 pKa = 9.67THH563 pKa = 6.09GVEE566 pKa = 3.73INADD570 pKa = 3.55VKK572 pKa = 10.8EE573 pKa = 4.55VIAKK577 pKa = 9.38QGNAIIKK584 pKa = 9.78NLYY587 pKa = 9.76SLYY590 pKa = 8.56KK591 pKa = 8.15TCVVVDD597 pKa = 5.01FTRR600 pKa = 11.84GLLEE604 pKa = 4.31FIGRR608 pKa = 11.84GGSANKK614 pKa = 10.01HH615 pKa = 5.74GLDD618 pKa = 3.36EE619 pKa = 4.29TDD621 pKa = 3.84IDD623 pKa = 4.65SEE625 pKa = 4.55TLNVIYY631 pKa = 10.73NHH633 pKa = 6.81IKK635 pKa = 10.31PSLTSNLNAYY645 pKa = 7.51QTTINKK651 pKa = 9.63IIEE654 pKa = 4.37KK655 pKa = 10.37VSGEE659 pKa = 4.17LGVQSEE665 pKa = 4.55EE666 pKa = 4.08LEE668 pKa = 4.25AANEE672 pKa = 4.04EE673 pKa = 4.52LFF675 pKa = 4.46
Molecular weight: 75.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
675 |
675 |
675 |
675.0 |
75.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.963 ± 0.0 | 1.481 ± 0.0 |
6.074 ± 0.0 | 6.37 ± 0.0 |
4.296 ± 0.0 | 5.037 ± 0.0 |
1.63 ± 0.0 | 8.148 ± 0.0 |
8.148 ± 0.0 | 8.741 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.926 ± 0.0 | 7.704 ± 0.0 |
3.407 ± 0.0 | 3.111 ± 0.0 |
2.667 ± 0.0 | 5.926 ± 0.0 |
6.815 ± 0.0 | 6.519 ± 0.0 |
0.741 ± 0.0 | 4.296 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
