
Aspergillus foetidus slow virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus
Average proteome isoelectric point is 7.17
Get precalculated fractions of proteins
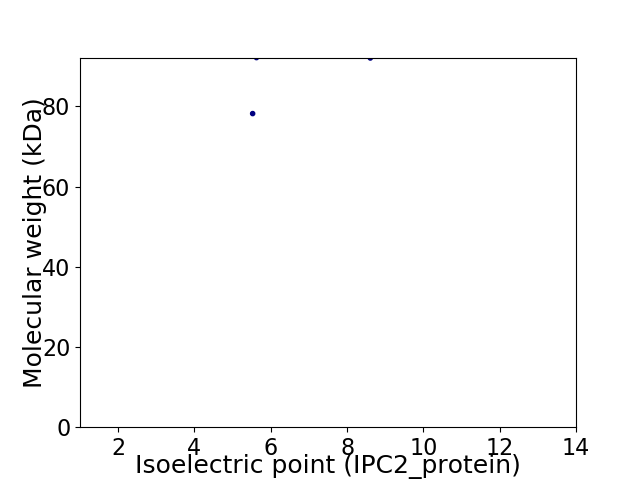
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
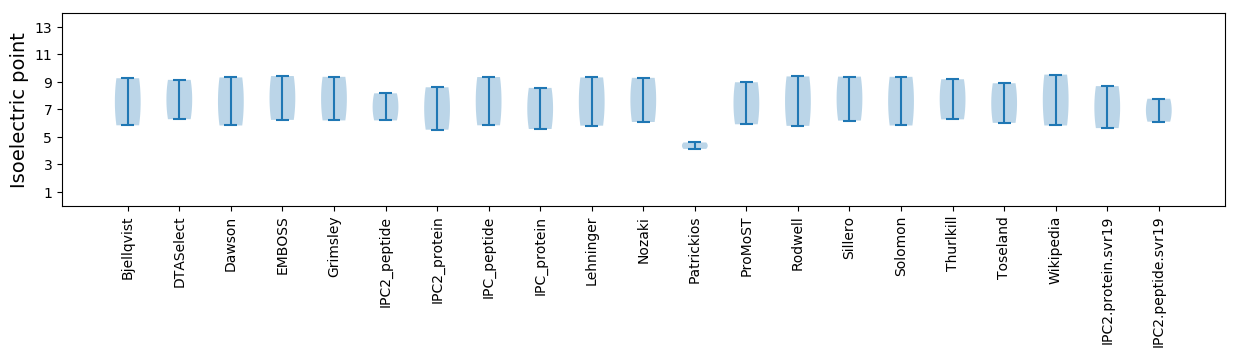
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J3JSF7|J3JSF7_9VIRU Coat protein OS=Aspergillus foetidus slow virus 1 OX=1087070 GN=CP PE=4 SV=1
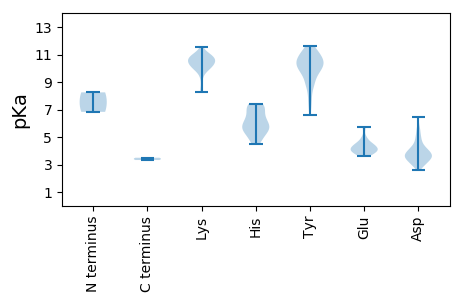
MM1 pKa = 6.83STIVRR6 pKa = 11.84NGYY9 pKa = 8.85LASVLANPRR18 pKa = 11.84GTRR21 pKa = 11.84INGDD25 pKa = 3.26AQFRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 9.05RR32 pKa = 11.84ANIRR36 pKa = 11.84TEE38 pKa = 3.88ATVGGVNDD46 pKa = 3.58ARR48 pKa = 11.84HH49 pKa = 5.2VQVSYY54 pKa = 10.85EE55 pKa = 3.88VGRR58 pKa = 11.84VHH60 pKa = 6.13RR61 pKa = 11.84TKK63 pKa = 10.88GAALAAAADD72 pKa = 3.56NLVRR76 pKa = 11.84VEE78 pKa = 4.04AAYY81 pKa = 7.63PTADD85 pKa = 3.23TLVEE89 pKa = 4.22DD90 pKa = 5.84FIGLAKK96 pKa = 10.29KK97 pKa = 10.42YY98 pKa = 10.34SNFSASFEE106 pKa = 4.18YY107 pKa = 10.81SSLAGVAEE115 pKa = 4.24RR116 pKa = 11.84LAKK119 pKa = 10.38GLAASSIWDD128 pKa = 4.02DD129 pKa = 3.43MTATDD134 pKa = 4.17LRR136 pKa = 11.84GNRR139 pKa = 11.84VLNISVLGTHH149 pKa = 7.29DD150 pKa = 4.04GPVNSLTNAVFIPRR164 pKa = 11.84LVNTAVTGDD173 pKa = 3.7VFTVMINAVAGEE185 pKa = 4.48GSAVVTDD192 pKa = 3.89VLEE195 pKa = 4.36LDD197 pKa = 3.52AATRR201 pKa = 11.84QPIVATVDD209 pKa = 2.95ADD211 pKa = 3.83GFSRR215 pKa = 11.84ACVEE219 pKa = 3.8ALRR222 pKa = 11.84IIGANMLAADD232 pKa = 4.09QGPLFAYY239 pKa = 9.8AVTRR243 pKa = 11.84GIHH246 pKa = 5.5NVFTVVGHH254 pKa = 5.33TDD256 pKa = 2.98EE257 pKa = 5.75GGVSRR262 pKa = 11.84DD263 pKa = 3.01WLRR266 pKa = 11.84ASGFAPPFGGIHH278 pKa = 6.27YY279 pKa = 7.88GLPVYY284 pKa = 10.65SGLPALSSTAPADD297 pKa = 3.16VSAYY301 pKa = 9.77VDD303 pKa = 4.76CIALTSAALVAHH315 pKa = 7.14SDD317 pKa = 3.39PGITYY322 pKa = 10.0DD323 pKa = 4.59DD324 pKa = 2.93RR325 pKa = 11.84WYY327 pKa = 10.14PSFYY331 pKa = 10.25QGTGADD337 pKa = 3.6TAEE340 pKa = 4.16VRR342 pKa = 11.84PGQHH346 pKa = 5.95QDD348 pKa = 2.59GTADD352 pKa = 3.07MGRR355 pKa = 11.84RR356 pKa = 11.84NWGQYY361 pKa = 6.63TSNMPRR367 pKa = 11.84FLDD370 pKa = 3.78YY371 pKa = 11.09YY372 pKa = 11.19LPGLGKK378 pKa = 10.7LFGADD383 pKa = 2.97GSVQLACTVARR394 pKa = 11.84THH396 pKa = 6.41SGLMPISRR404 pKa = 11.84HH405 pKa = 4.85FNYY408 pKa = 10.61ASIAPFFWIEE418 pKa = 3.85PTSLIPHH425 pKa = 7.28DD426 pKa = 4.36FLGSPAEE433 pKa = 4.14LNGSGSYY440 pKa = 9.09CTVDD444 pKa = 3.5VPRR447 pKa = 11.84EE448 pKa = 3.7KK449 pKa = 10.88AGFEE453 pKa = 5.03DD454 pKa = 3.5ITAAGAADD462 pKa = 3.81VAYY465 pKa = 10.29SAYY468 pKa = 10.04NVLMRR473 pKa = 11.84NPRR476 pKa = 11.84PAWFFAHH483 pKa = 7.21WLNHH487 pKa = 5.67PLNGLGAISVRR498 pKa = 11.84QLDD501 pKa = 4.14PNGIIHH507 pKa = 7.41PGACPDD513 pKa = 3.74HH514 pKa = 7.23PDD516 pKa = 2.97VRR518 pKa = 11.84DD519 pKa = 3.4RR520 pKa = 11.84VEE522 pKa = 5.07AALPWTSFMWTRR534 pKa = 11.84GQSPFPAPGEE544 pKa = 3.86LLNLSGTAGFMVKK557 pKa = 10.08HH558 pKa = 5.67LTFDD562 pKa = 4.23DD563 pKa = 4.32DD564 pKa = 5.61AIPTPEE570 pKa = 4.37HH571 pKa = 6.7LPTARR576 pKa = 11.84EE577 pKa = 3.9FEE579 pKa = 4.44GVSVRR584 pKa = 11.84ITCGRR589 pKa = 11.84PMSIANGTSNSPDD602 pKa = 2.9STARR606 pKa = 11.84RR607 pKa = 11.84ARR609 pKa = 11.84TRR611 pKa = 11.84AARR614 pKa = 11.84EE615 pKa = 3.65LAAATARR622 pKa = 11.84DD623 pKa = 3.7RR624 pKa = 11.84LFGRR628 pKa = 11.84PAVAEE633 pKa = 4.1MPILTTAPVMRR644 pKa = 11.84TKK646 pKa = 10.6VAAPPPQEE654 pKa = 4.23PPQVSGGVSGWSRR667 pKa = 11.84AVPAAEE673 pKa = 4.36NGVNDD678 pKa = 4.75DD679 pKa = 4.51RR680 pKa = 11.84NPDD683 pKa = 3.34GAAVPVVPHH692 pKa = 5.66YY693 pKa = 10.95NSXKK697 pKa = 9.7FPVVARR703 pKa = 11.84PQAGAVIGGGAAPAAPGAPAADD725 pKa = 4.28EE726 pKa = 5.59DD727 pKa = 5.37DD728 pKa = 4.22NQPPQPVAIGNXNN741 pKa = 3.33
MM1 pKa = 6.83STIVRR6 pKa = 11.84NGYY9 pKa = 8.85LASVLANPRR18 pKa = 11.84GTRR21 pKa = 11.84INGDD25 pKa = 3.26AQFRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 9.05RR32 pKa = 11.84ANIRR36 pKa = 11.84TEE38 pKa = 3.88ATVGGVNDD46 pKa = 3.58ARR48 pKa = 11.84HH49 pKa = 5.2VQVSYY54 pKa = 10.85EE55 pKa = 3.88VGRR58 pKa = 11.84VHH60 pKa = 6.13RR61 pKa = 11.84TKK63 pKa = 10.88GAALAAAADD72 pKa = 3.56NLVRR76 pKa = 11.84VEE78 pKa = 4.04AAYY81 pKa = 7.63PTADD85 pKa = 3.23TLVEE89 pKa = 4.22DD90 pKa = 5.84FIGLAKK96 pKa = 10.29KK97 pKa = 10.42YY98 pKa = 10.34SNFSASFEE106 pKa = 4.18YY107 pKa = 10.81SSLAGVAEE115 pKa = 4.24RR116 pKa = 11.84LAKK119 pKa = 10.38GLAASSIWDD128 pKa = 4.02DD129 pKa = 3.43MTATDD134 pKa = 4.17LRR136 pKa = 11.84GNRR139 pKa = 11.84VLNISVLGTHH149 pKa = 7.29DD150 pKa = 4.04GPVNSLTNAVFIPRR164 pKa = 11.84LVNTAVTGDD173 pKa = 3.7VFTVMINAVAGEE185 pKa = 4.48GSAVVTDD192 pKa = 3.89VLEE195 pKa = 4.36LDD197 pKa = 3.52AATRR201 pKa = 11.84QPIVATVDD209 pKa = 2.95ADD211 pKa = 3.83GFSRR215 pKa = 11.84ACVEE219 pKa = 3.8ALRR222 pKa = 11.84IIGANMLAADD232 pKa = 4.09QGPLFAYY239 pKa = 9.8AVTRR243 pKa = 11.84GIHH246 pKa = 5.5NVFTVVGHH254 pKa = 5.33TDD256 pKa = 2.98EE257 pKa = 5.75GGVSRR262 pKa = 11.84DD263 pKa = 3.01WLRR266 pKa = 11.84ASGFAPPFGGIHH278 pKa = 6.27YY279 pKa = 7.88GLPVYY284 pKa = 10.65SGLPALSSTAPADD297 pKa = 3.16VSAYY301 pKa = 9.77VDD303 pKa = 4.76CIALTSAALVAHH315 pKa = 7.14SDD317 pKa = 3.39PGITYY322 pKa = 10.0DD323 pKa = 4.59DD324 pKa = 2.93RR325 pKa = 11.84WYY327 pKa = 10.14PSFYY331 pKa = 10.25QGTGADD337 pKa = 3.6TAEE340 pKa = 4.16VRR342 pKa = 11.84PGQHH346 pKa = 5.95QDD348 pKa = 2.59GTADD352 pKa = 3.07MGRR355 pKa = 11.84RR356 pKa = 11.84NWGQYY361 pKa = 6.63TSNMPRR367 pKa = 11.84FLDD370 pKa = 3.78YY371 pKa = 11.09YY372 pKa = 11.19LPGLGKK378 pKa = 10.7LFGADD383 pKa = 2.97GSVQLACTVARR394 pKa = 11.84THH396 pKa = 6.41SGLMPISRR404 pKa = 11.84HH405 pKa = 4.85FNYY408 pKa = 10.61ASIAPFFWIEE418 pKa = 3.85PTSLIPHH425 pKa = 7.28DD426 pKa = 4.36FLGSPAEE433 pKa = 4.14LNGSGSYY440 pKa = 9.09CTVDD444 pKa = 3.5VPRR447 pKa = 11.84EE448 pKa = 3.7KK449 pKa = 10.88AGFEE453 pKa = 5.03DD454 pKa = 3.5ITAAGAADD462 pKa = 3.81VAYY465 pKa = 10.29SAYY468 pKa = 10.04NVLMRR473 pKa = 11.84NPRR476 pKa = 11.84PAWFFAHH483 pKa = 7.21WLNHH487 pKa = 5.67PLNGLGAISVRR498 pKa = 11.84QLDD501 pKa = 4.14PNGIIHH507 pKa = 7.41PGACPDD513 pKa = 3.74HH514 pKa = 7.23PDD516 pKa = 2.97VRR518 pKa = 11.84DD519 pKa = 3.4RR520 pKa = 11.84VEE522 pKa = 5.07AALPWTSFMWTRR534 pKa = 11.84GQSPFPAPGEE544 pKa = 3.86LLNLSGTAGFMVKK557 pKa = 10.08HH558 pKa = 5.67LTFDD562 pKa = 4.23DD563 pKa = 4.32DD564 pKa = 5.61AIPTPEE570 pKa = 4.37HH571 pKa = 6.7LPTARR576 pKa = 11.84EE577 pKa = 3.9FEE579 pKa = 4.44GVSVRR584 pKa = 11.84ITCGRR589 pKa = 11.84PMSIANGTSNSPDD602 pKa = 2.9STARR606 pKa = 11.84RR607 pKa = 11.84ARR609 pKa = 11.84TRR611 pKa = 11.84AARR614 pKa = 11.84EE615 pKa = 3.65LAAATARR622 pKa = 11.84DD623 pKa = 3.7RR624 pKa = 11.84LFGRR628 pKa = 11.84PAVAEE633 pKa = 4.1MPILTTAPVMRR644 pKa = 11.84TKK646 pKa = 10.6VAAPPPQEE654 pKa = 4.23PPQVSGGVSGWSRR667 pKa = 11.84AVPAAEE673 pKa = 4.36NGVNDD678 pKa = 4.75DD679 pKa = 4.51RR680 pKa = 11.84NPDD683 pKa = 3.34GAAVPVVPHH692 pKa = 5.66YY693 pKa = 10.95NSXKK697 pKa = 9.7FPVVARR703 pKa = 11.84PQAGAVIGGGAAPAAPGAPAADD725 pKa = 4.28EE726 pKa = 5.59DD727 pKa = 5.37DD728 pKa = 4.22NQPPQPVAIGNXNN741 pKa = 3.33
Molecular weight: 78.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J3JSF7|J3JSF7_9VIRU Coat protein OS=Aspergillus foetidus slow virus 1 OX=1087070 GN=CP PE=4 SV=1
MM1 pKa = 8.25DD2 pKa = 3.63IQTRR6 pKa = 11.84ISEE9 pKa = 4.48FGRR12 pKa = 11.84IGTYY16 pKa = 10.6LFGLINGTDD25 pKa = 3.89FMDD28 pKa = 4.05QINSHH33 pKa = 5.07GTYY36 pKa = 8.99VAKK39 pKa = 10.92LIALRR44 pKa = 11.84NSMSSLRR51 pKa = 11.84EE52 pKa = 3.65RR53 pKa = 11.84HH54 pKa = 5.89ALLPAAASLLLLPFPLQVEE73 pKa = 4.12LSIGDD78 pKa = 3.6VLLLLRR84 pKa = 11.84SAFNLSYY91 pKa = 9.96GTTIQAAISSNAIPEE106 pKa = 4.12ILSDD110 pKa = 3.77NLAGRR115 pKa = 11.84ARR117 pKa = 11.84RR118 pKa = 11.84IVTLVTQRR126 pKa = 11.84PDD128 pKa = 2.98FRR130 pKa = 11.84EE131 pKa = 3.84KK132 pKa = 9.96YY133 pKa = 10.25FPVKK137 pKa = 10.03KK138 pKa = 10.21HH139 pKa = 6.32PGAQTKK145 pKa = 10.94ANVHH149 pKa = 5.6LAGLVRR155 pKa = 11.84SAVVLHH161 pKa = 5.87GRR163 pKa = 11.84GIIGRR168 pKa = 11.84VLLATAGRR176 pKa = 11.84LTDD179 pKa = 4.13DD180 pKa = 3.6QASSLLIYY188 pKa = 10.69GSGLLPTFGALGWAIATHH206 pKa = 5.4MVLYY210 pKa = 10.29PDD212 pKa = 4.99DD213 pKa = 4.9AKK215 pKa = 11.51ALNGALKK222 pKa = 10.76ALGTNSTPYY231 pKa = 10.81GCLLVEE237 pKa = 4.61ANTLQGRR244 pKa = 11.84GVGAIDD250 pKa = 3.4MRR252 pKa = 11.84EE253 pKa = 3.98KK254 pKa = 10.78ARR256 pKa = 11.84EE257 pKa = 3.76RR258 pKa = 11.84CDD260 pKa = 3.31PDD262 pKa = 5.56FIQKK266 pKa = 10.19SVVSANFDD274 pKa = 3.47EE275 pKa = 4.89LRR277 pKa = 11.84EE278 pKa = 4.0HH279 pKa = 5.89VRR281 pKa = 11.84AVITTEE287 pKa = 3.65LSGRR291 pKa = 11.84DD292 pKa = 3.28VKK294 pKa = 10.9MPTLSDD300 pKa = 3.59FWTSRR305 pKa = 11.84WAWCVNGSEE314 pKa = 4.34TAKK317 pKa = 10.46SDD319 pKa = 3.56EE320 pKa = 4.43VLGLDD325 pKa = 3.38PKK327 pKa = 10.76AYY329 pKa = 9.77KK330 pKa = 9.16RR331 pKa = 11.84THH333 pKa = 4.52TRR335 pKa = 11.84SYY337 pKa = 10.77RR338 pKa = 11.84RR339 pKa = 11.84MAAEE343 pKa = 4.07SVRR346 pKa = 11.84DD347 pKa = 3.88EE348 pKa = 5.72PISTWDD354 pKa = 3.11GTTYY358 pKa = 11.35ASASPKK364 pKa = 10.55LEE366 pKa = 4.16AGKK369 pKa = 8.26TRR371 pKa = 11.84AIFACDD377 pKa = 3.1TRR379 pKa = 11.84SYY381 pKa = 11.04FAFSWCLNAVQKK393 pKa = 10.68AWRR396 pKa = 11.84NSRR399 pKa = 11.84VLLDD403 pKa = 3.95PGIGGHH409 pKa = 6.59LGMVNRR415 pKa = 11.84IRR417 pKa = 11.84HH418 pKa = 4.75AQRR421 pKa = 11.84GGGVNLMLDD430 pKa = 3.56FDD432 pKa = 5.04DD433 pKa = 6.45FNSQHH438 pKa = 6.08HH439 pKa = 5.44TVAMQIVFDD448 pKa = 4.56EE449 pKa = 4.34LCKK452 pKa = 11.12YY453 pKa = 10.23IGAPDD458 pKa = 3.97WYY460 pKa = 10.52RR461 pKa = 11.84HH462 pKa = 4.54VLVNSFTRR470 pKa = 11.84THH472 pKa = 5.77VSIDD476 pKa = 3.51GRR478 pKa = 11.84WEE480 pKa = 3.99LVQGTLMSGHH490 pKa = 7.17RR491 pKa = 11.84GTTFINSVLNAAYY504 pKa = 9.68IRR506 pKa = 11.84MAVGGPYY513 pKa = 9.75FDD515 pKa = 5.12SLLSLHH521 pKa = 6.57TGDD524 pKa = 5.19DD525 pKa = 3.71VYY527 pKa = 11.3IRR529 pKa = 11.84ANTLGDD535 pKa = 3.52CDD537 pKa = 4.69YY538 pKa = 11.25ILNRR542 pKa = 11.84CRR544 pKa = 11.84DD545 pKa = 3.67YY546 pKa = 11.61GCRR549 pKa = 11.84LNPAKK554 pKa = 10.49QSVGYY559 pKa = 10.18YY560 pKa = 8.92GAEE563 pKa = 4.13FLRR566 pKa = 11.84VAIRR570 pKa = 11.84GEE572 pKa = 3.64RR573 pKa = 11.84AYY575 pKa = 10.46GYY577 pKa = 8.7FARR580 pKa = 11.84GVSGFVNGNWTSSDD594 pKa = 3.71PLSLTEE600 pKa = 4.57GLSSAIASCRR610 pKa = 11.84TLINRR615 pKa = 11.84SGDD618 pKa = 3.22ASLADD623 pKa = 4.08LLGPAIRR630 pKa = 11.84YY631 pKa = 8.51RR632 pKa = 11.84RR633 pKa = 11.84GLTTRR638 pKa = 11.84QTIQLLRR645 pKa = 11.84GTLSLEE651 pKa = 3.89GSPVFNTNYY660 pKa = 9.84IIRR663 pKa = 11.84NLRR666 pKa = 11.84VEE668 pKa = 4.62GVDD671 pKa = 3.84KK672 pKa = 10.34EE673 pKa = 4.62TVPIDD678 pKa = 3.89NRR680 pKa = 11.84WKK682 pKa = 10.38RR683 pKa = 11.84YY684 pKa = 7.36ATTDD688 pKa = 3.39YY689 pKa = 10.47LTDD692 pKa = 3.47HH693 pKa = 7.24LSPVEE698 pKa = 3.95VEE700 pKa = 4.8AIHH703 pKa = 6.76LAKK706 pKa = 9.91TDD708 pKa = 4.27PGPLMIASGYY718 pKa = 10.34RR719 pKa = 11.84KK720 pKa = 10.07GLNLDD725 pKa = 4.0DD726 pKa = 5.57KK727 pKa = 10.22GTPSQVRR734 pKa = 11.84FRR736 pKa = 11.84PLPPRR741 pKa = 11.84LATGFASASQLTARR755 pKa = 11.84RR756 pKa = 11.84AEE758 pKa = 4.19PGCLAKK764 pKa = 10.99YY765 pKa = 9.5PVLSLIAGRR774 pKa = 11.84LTDD777 pKa = 4.18DD778 pKa = 3.77EE779 pKa = 4.82LRR781 pKa = 11.84HH782 pKa = 5.24LVAMEE787 pKa = 4.56GGNNTTSDD795 pKa = 2.93IRR797 pKa = 11.84KK798 pKa = 8.37EE799 pKa = 4.06AFGEE803 pKa = 4.41SPTSKK808 pKa = 10.7NIIGFLSYY816 pKa = 11.38ADD818 pKa = 3.72AAALSKK824 pKa = 9.77VTSSGNIFTSISVRR838 pKa = 11.84VV839 pKa = 3.49
MM1 pKa = 8.25DD2 pKa = 3.63IQTRR6 pKa = 11.84ISEE9 pKa = 4.48FGRR12 pKa = 11.84IGTYY16 pKa = 10.6LFGLINGTDD25 pKa = 3.89FMDD28 pKa = 4.05QINSHH33 pKa = 5.07GTYY36 pKa = 8.99VAKK39 pKa = 10.92LIALRR44 pKa = 11.84NSMSSLRR51 pKa = 11.84EE52 pKa = 3.65RR53 pKa = 11.84HH54 pKa = 5.89ALLPAAASLLLLPFPLQVEE73 pKa = 4.12LSIGDD78 pKa = 3.6VLLLLRR84 pKa = 11.84SAFNLSYY91 pKa = 9.96GTTIQAAISSNAIPEE106 pKa = 4.12ILSDD110 pKa = 3.77NLAGRR115 pKa = 11.84ARR117 pKa = 11.84RR118 pKa = 11.84IVTLVTQRR126 pKa = 11.84PDD128 pKa = 2.98FRR130 pKa = 11.84EE131 pKa = 3.84KK132 pKa = 9.96YY133 pKa = 10.25FPVKK137 pKa = 10.03KK138 pKa = 10.21HH139 pKa = 6.32PGAQTKK145 pKa = 10.94ANVHH149 pKa = 5.6LAGLVRR155 pKa = 11.84SAVVLHH161 pKa = 5.87GRR163 pKa = 11.84GIIGRR168 pKa = 11.84VLLATAGRR176 pKa = 11.84LTDD179 pKa = 4.13DD180 pKa = 3.6QASSLLIYY188 pKa = 10.69GSGLLPTFGALGWAIATHH206 pKa = 5.4MVLYY210 pKa = 10.29PDD212 pKa = 4.99DD213 pKa = 4.9AKK215 pKa = 11.51ALNGALKK222 pKa = 10.76ALGTNSTPYY231 pKa = 10.81GCLLVEE237 pKa = 4.61ANTLQGRR244 pKa = 11.84GVGAIDD250 pKa = 3.4MRR252 pKa = 11.84EE253 pKa = 3.98KK254 pKa = 10.78ARR256 pKa = 11.84EE257 pKa = 3.76RR258 pKa = 11.84CDD260 pKa = 3.31PDD262 pKa = 5.56FIQKK266 pKa = 10.19SVVSANFDD274 pKa = 3.47EE275 pKa = 4.89LRR277 pKa = 11.84EE278 pKa = 4.0HH279 pKa = 5.89VRR281 pKa = 11.84AVITTEE287 pKa = 3.65LSGRR291 pKa = 11.84DD292 pKa = 3.28VKK294 pKa = 10.9MPTLSDD300 pKa = 3.59FWTSRR305 pKa = 11.84WAWCVNGSEE314 pKa = 4.34TAKK317 pKa = 10.46SDD319 pKa = 3.56EE320 pKa = 4.43VLGLDD325 pKa = 3.38PKK327 pKa = 10.76AYY329 pKa = 9.77KK330 pKa = 9.16RR331 pKa = 11.84THH333 pKa = 4.52TRR335 pKa = 11.84SYY337 pKa = 10.77RR338 pKa = 11.84RR339 pKa = 11.84MAAEE343 pKa = 4.07SVRR346 pKa = 11.84DD347 pKa = 3.88EE348 pKa = 5.72PISTWDD354 pKa = 3.11GTTYY358 pKa = 11.35ASASPKK364 pKa = 10.55LEE366 pKa = 4.16AGKK369 pKa = 8.26TRR371 pKa = 11.84AIFACDD377 pKa = 3.1TRR379 pKa = 11.84SYY381 pKa = 11.04FAFSWCLNAVQKK393 pKa = 10.68AWRR396 pKa = 11.84NSRR399 pKa = 11.84VLLDD403 pKa = 3.95PGIGGHH409 pKa = 6.59LGMVNRR415 pKa = 11.84IRR417 pKa = 11.84HH418 pKa = 4.75AQRR421 pKa = 11.84GGGVNLMLDD430 pKa = 3.56FDD432 pKa = 5.04DD433 pKa = 6.45FNSQHH438 pKa = 6.08HH439 pKa = 5.44TVAMQIVFDD448 pKa = 4.56EE449 pKa = 4.34LCKK452 pKa = 11.12YY453 pKa = 10.23IGAPDD458 pKa = 3.97WYY460 pKa = 10.52RR461 pKa = 11.84HH462 pKa = 4.54VLVNSFTRR470 pKa = 11.84THH472 pKa = 5.77VSIDD476 pKa = 3.51GRR478 pKa = 11.84WEE480 pKa = 3.99LVQGTLMSGHH490 pKa = 7.17RR491 pKa = 11.84GTTFINSVLNAAYY504 pKa = 9.68IRR506 pKa = 11.84MAVGGPYY513 pKa = 9.75FDD515 pKa = 5.12SLLSLHH521 pKa = 6.57TGDD524 pKa = 5.19DD525 pKa = 3.71VYY527 pKa = 11.3IRR529 pKa = 11.84ANTLGDD535 pKa = 3.52CDD537 pKa = 4.69YY538 pKa = 11.25ILNRR542 pKa = 11.84CRR544 pKa = 11.84DD545 pKa = 3.67YY546 pKa = 11.61GCRR549 pKa = 11.84LNPAKK554 pKa = 10.49QSVGYY559 pKa = 10.18YY560 pKa = 8.92GAEE563 pKa = 4.13FLRR566 pKa = 11.84VAIRR570 pKa = 11.84GEE572 pKa = 3.64RR573 pKa = 11.84AYY575 pKa = 10.46GYY577 pKa = 8.7FARR580 pKa = 11.84GVSGFVNGNWTSSDD594 pKa = 3.71PLSLTEE600 pKa = 4.57GLSSAIASCRR610 pKa = 11.84TLINRR615 pKa = 11.84SGDD618 pKa = 3.22ASLADD623 pKa = 4.08LLGPAIRR630 pKa = 11.84YY631 pKa = 8.51RR632 pKa = 11.84RR633 pKa = 11.84GLTTRR638 pKa = 11.84QTIQLLRR645 pKa = 11.84GTLSLEE651 pKa = 3.89GSPVFNTNYY660 pKa = 9.84IIRR663 pKa = 11.84NLRR666 pKa = 11.84VEE668 pKa = 4.62GVDD671 pKa = 3.84KK672 pKa = 10.34EE673 pKa = 4.62TVPIDD678 pKa = 3.89NRR680 pKa = 11.84WKK682 pKa = 10.38RR683 pKa = 11.84YY684 pKa = 7.36ATTDD688 pKa = 3.39YY689 pKa = 10.47LTDD692 pKa = 3.47HH693 pKa = 7.24LSPVEE698 pKa = 3.95VEE700 pKa = 4.8AIHH703 pKa = 6.76LAKK706 pKa = 9.91TDD708 pKa = 4.27PGPLMIASGYY718 pKa = 10.34RR719 pKa = 11.84KK720 pKa = 10.07GLNLDD725 pKa = 4.0DD726 pKa = 5.57KK727 pKa = 10.22GTPSQVRR734 pKa = 11.84FRR736 pKa = 11.84PLPPRR741 pKa = 11.84LATGFASASQLTARR755 pKa = 11.84RR756 pKa = 11.84AEE758 pKa = 4.19PGCLAKK764 pKa = 10.99YY765 pKa = 9.5PVLSLIAGRR774 pKa = 11.84LTDD777 pKa = 4.18DD778 pKa = 3.77EE779 pKa = 4.82LRR781 pKa = 11.84HH782 pKa = 5.24LVAMEE787 pKa = 4.56GGNNTTSDD795 pKa = 2.93IRR797 pKa = 11.84KK798 pKa = 8.37EE799 pKa = 4.06AFGEE803 pKa = 4.41SPTSKK808 pKa = 10.7NIIGFLSYY816 pKa = 11.38ADD818 pKa = 3.72AAALSKK824 pKa = 9.77VTSSGNIFTSISVRR838 pKa = 11.84VV839 pKa = 3.49
Molecular weight: 92.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1580 |
741 |
839 |
790.0 |
85.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.582 ± 1.673 | 1.076 ± 0.172 |
5.949 ± 0.167 | 3.544 ± 0.11 |
3.608 ± 0.111 | 8.861 ± 0.204 |
2.342 ± 0.056 | 4.747 ± 0.539 |
2.278 ± 0.688 | 8.924 ± 1.582 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.709 ± 0.029 | 4.367 ± 0.143 |
5.759 ± 1.337 | 2.215 ± 0.036 |
7.532 ± 0.507 | 7.089 ± 0.569 |
6.582 ± 0.242 | 7.215 ± 0.832 |
1.329 ± 0.013 | 3.165 ± 0.214 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
