
Actinotalea fermentans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia<
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
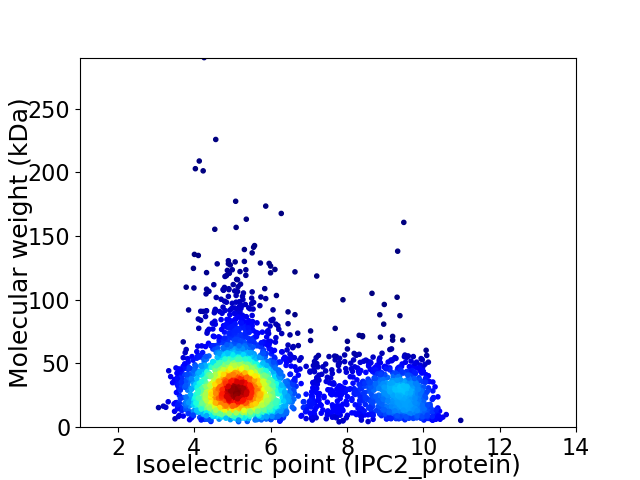
Virtual 2D-PAGE plot for 3422 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A511YV90|A0A511YV90_9CELL NAD(P)-dependent oxidoreductase OS=Actinotalea fermentans OX=43671 GN=AFE02nite_08560 PE=4 SV=1
MM1 pKa = 7.42AAVFSNRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84ALAMTAGTAAVVVALAGCGGADD32 pKa = 4.62DD33 pKa = 5.29GGSGDD38 pKa = 3.46GGGAGGEE45 pKa = 4.16LVIGTTDD52 pKa = 3.87KK53 pKa = 10.49ITTIDD58 pKa = 3.49PAGSYY63 pKa = 11.17DD64 pKa = 3.54NGSFAVMNQVFPFLLNTPLGSPDD87 pKa = 3.76VEE89 pKa = 3.9PDD91 pKa = 3.01IAEE94 pKa = 4.25SAEE97 pKa = 4.09FTSPTEE103 pKa = 3.78YY104 pKa = 10.4TVVLKK109 pKa = 10.98EE110 pKa = 4.02GLTFANGNEE119 pKa = 4.2LTSSDD124 pKa = 3.59VKK126 pKa = 10.49FTFDD130 pKa = 2.57RR131 pKa = 11.84MQAIFEE137 pKa = 4.3SGADD141 pKa = 3.58GGNGPGSLLYY151 pKa = 10.6NLEE154 pKa = 4.14STEE157 pKa = 4.08ATDD160 pKa = 3.2EE161 pKa = 4.24LTVVFHH167 pKa = 6.83LKK169 pKa = 10.4SEE171 pKa = 4.12NDD173 pKa = 3.26QVFPQILSSPAGPIVDD189 pKa = 3.9EE190 pKa = 5.08DD191 pKa = 4.14VFAADD196 pKa = 5.06ALTPDD201 pKa = 3.81ADD203 pKa = 3.48IVAGNAFAGPYY214 pKa = 10.27VITSYY219 pKa = 11.45DD220 pKa = 3.76LNNLISYY227 pKa = 7.56EE228 pKa = 4.27ANPDD232 pKa = 3.55YY233 pKa = 10.88QGLLGAPRR241 pKa = 11.84NSAVNVKK248 pKa = 10.08YY249 pKa = 10.55YY250 pKa = 11.3AEE252 pKa = 4.27ASNLKK257 pKa = 10.42LDD259 pKa = 3.89VQEE262 pKa = 4.8GNVDD266 pKa = 3.16VAYY269 pKa = 10.41RR270 pKa = 11.84SLSATDD276 pKa = 3.71IEE278 pKa = 4.98DD279 pKa = 3.5LRR281 pKa = 11.84GDD283 pKa = 3.78EE284 pKa = 4.15NVEE287 pKa = 4.21VVDD290 pKa = 4.75GPGGEE295 pKa = 3.55IRR297 pKa = 11.84YY298 pKa = 9.34IVFNFDD304 pKa = 3.41TQPYY308 pKa = 9.09GATTAEE314 pKa = 3.98ADD316 pKa = 3.66EE317 pKa = 4.68AKK319 pKa = 10.75ALAVRR324 pKa = 11.84QAVAHH329 pKa = 6.5LIDD332 pKa = 3.83RR333 pKa = 11.84AEE335 pKa = 4.27LAEE338 pKa = 3.93QVYY341 pKa = 10.4KK342 pKa = 10.16GTYY345 pKa = 7.21TPLYY349 pKa = 10.19SFVPDD354 pKa = 3.66GLTGATEE361 pKa = 4.06ALKK364 pKa = 10.63DD365 pKa = 3.96LYY367 pKa = 11.27GDD369 pKa = 3.84GDD371 pKa = 4.05GGPDD375 pKa = 3.43ADD377 pKa = 3.77AAAAVLEE384 pKa = 4.49EE385 pKa = 4.74AGVTTPVQLSLQYY398 pKa = 11.43SNDD401 pKa = 3.17HH402 pKa = 6.35YY403 pKa = 11.31GPSSGDD409 pKa = 3.07EE410 pKa = 3.82YY411 pKa = 11.92ALIKK415 pKa = 10.69DD416 pKa = 3.71QLEE419 pKa = 4.2SSGLFTVDD427 pKa = 3.29LQTTEE432 pKa = 3.92WVQYY436 pKa = 10.29AQDD439 pKa = 3.85RR440 pKa = 11.84TSDD443 pKa = 3.69VYY445 pKa = 10.52PAYY448 pKa = 10.27QLGWFPDD455 pKa = 3.67YY456 pKa = 11.48SDD458 pKa = 4.11ADD460 pKa = 3.57NYY462 pKa = 8.91LTPFFLVEE470 pKa = 4.03NFLGNHH476 pKa = 5.49YY477 pKa = 10.88VNQEE481 pKa = 3.95VNDD484 pKa = 4.43MILEE488 pKa = 4.18QAVTTDD494 pKa = 2.83ADD496 pKa = 3.32ARR498 pKa = 11.84TALIEE503 pKa = 4.93DD504 pKa = 3.59IQAAVAQDD512 pKa = 4.35LSTVPYY518 pKa = 10.3LQGAQVAVVRR528 pKa = 11.84TGVTGAGDD536 pKa = 3.48TLDD539 pKa = 3.92ASFKK543 pKa = 10.38FRR545 pKa = 11.84YY546 pKa = 8.91GALAKK551 pKa = 10.5DD552 pKa = 3.61
MM1 pKa = 7.42AAVFSNRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84ALAMTAGTAAVVVALAGCGGADD32 pKa = 4.62DD33 pKa = 5.29GGSGDD38 pKa = 3.46GGGAGGEE45 pKa = 4.16LVIGTTDD52 pKa = 3.87KK53 pKa = 10.49ITTIDD58 pKa = 3.49PAGSYY63 pKa = 11.17DD64 pKa = 3.54NGSFAVMNQVFPFLLNTPLGSPDD87 pKa = 3.76VEE89 pKa = 3.9PDD91 pKa = 3.01IAEE94 pKa = 4.25SAEE97 pKa = 4.09FTSPTEE103 pKa = 3.78YY104 pKa = 10.4TVVLKK109 pKa = 10.98EE110 pKa = 4.02GLTFANGNEE119 pKa = 4.2LTSSDD124 pKa = 3.59VKK126 pKa = 10.49FTFDD130 pKa = 2.57RR131 pKa = 11.84MQAIFEE137 pKa = 4.3SGADD141 pKa = 3.58GGNGPGSLLYY151 pKa = 10.6NLEE154 pKa = 4.14STEE157 pKa = 4.08ATDD160 pKa = 3.2EE161 pKa = 4.24LTVVFHH167 pKa = 6.83LKK169 pKa = 10.4SEE171 pKa = 4.12NDD173 pKa = 3.26QVFPQILSSPAGPIVDD189 pKa = 3.9EE190 pKa = 5.08DD191 pKa = 4.14VFAADD196 pKa = 5.06ALTPDD201 pKa = 3.81ADD203 pKa = 3.48IVAGNAFAGPYY214 pKa = 10.27VITSYY219 pKa = 11.45DD220 pKa = 3.76LNNLISYY227 pKa = 7.56EE228 pKa = 4.27ANPDD232 pKa = 3.55YY233 pKa = 10.88QGLLGAPRR241 pKa = 11.84NSAVNVKK248 pKa = 10.08YY249 pKa = 10.55YY250 pKa = 11.3AEE252 pKa = 4.27ASNLKK257 pKa = 10.42LDD259 pKa = 3.89VQEE262 pKa = 4.8GNVDD266 pKa = 3.16VAYY269 pKa = 10.41RR270 pKa = 11.84SLSATDD276 pKa = 3.71IEE278 pKa = 4.98DD279 pKa = 3.5LRR281 pKa = 11.84GDD283 pKa = 3.78EE284 pKa = 4.15NVEE287 pKa = 4.21VVDD290 pKa = 4.75GPGGEE295 pKa = 3.55IRR297 pKa = 11.84YY298 pKa = 9.34IVFNFDD304 pKa = 3.41TQPYY308 pKa = 9.09GATTAEE314 pKa = 3.98ADD316 pKa = 3.66EE317 pKa = 4.68AKK319 pKa = 10.75ALAVRR324 pKa = 11.84QAVAHH329 pKa = 6.5LIDD332 pKa = 3.83RR333 pKa = 11.84AEE335 pKa = 4.27LAEE338 pKa = 3.93QVYY341 pKa = 10.4KK342 pKa = 10.16GTYY345 pKa = 7.21TPLYY349 pKa = 10.19SFVPDD354 pKa = 3.66GLTGATEE361 pKa = 4.06ALKK364 pKa = 10.63DD365 pKa = 3.96LYY367 pKa = 11.27GDD369 pKa = 3.84GDD371 pKa = 4.05GGPDD375 pKa = 3.43ADD377 pKa = 3.77AAAAVLEE384 pKa = 4.49EE385 pKa = 4.74AGVTTPVQLSLQYY398 pKa = 11.43SNDD401 pKa = 3.17HH402 pKa = 6.35YY403 pKa = 11.31GPSSGDD409 pKa = 3.07EE410 pKa = 3.82YY411 pKa = 11.92ALIKK415 pKa = 10.69DD416 pKa = 3.71QLEE419 pKa = 4.2SSGLFTVDD427 pKa = 3.29LQTTEE432 pKa = 3.92WVQYY436 pKa = 10.29AQDD439 pKa = 3.85RR440 pKa = 11.84TSDD443 pKa = 3.69VYY445 pKa = 10.52PAYY448 pKa = 10.27QLGWFPDD455 pKa = 3.67YY456 pKa = 11.48SDD458 pKa = 4.11ADD460 pKa = 3.57NYY462 pKa = 8.91LTPFFLVEE470 pKa = 4.03NFLGNHH476 pKa = 5.49YY477 pKa = 10.88VNQEE481 pKa = 3.95VNDD484 pKa = 4.43MILEE488 pKa = 4.18QAVTTDD494 pKa = 2.83ADD496 pKa = 3.32ARR498 pKa = 11.84TALIEE503 pKa = 4.93DD504 pKa = 3.59IQAAVAQDD512 pKa = 4.35LSTVPYY518 pKa = 10.3LQGAQVAVVRR528 pKa = 11.84TGVTGAGDD536 pKa = 3.48TLDD539 pKa = 3.92ASFKK543 pKa = 10.38FRR545 pKa = 11.84YY546 pKa = 8.91GALAKK551 pKa = 10.5DD552 pKa = 3.61
Molecular weight: 58.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A511Z254|A0A511Z254_9CELL Uncharacterized protein OS=Actinotalea fermentans OX=43671 GN=AFE02nite_32100 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.88GRR40 pKa = 11.84EE41 pKa = 3.56NLSAA45 pKa = 4.59
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.88GRR40 pKa = 11.84EE41 pKa = 3.56NLSAA45 pKa = 4.59
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1139067 |
39 |
2802 |
332.9 |
35.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.222 ± 0.055 | 0.599 ± 0.01 |
6.273 ± 0.033 | 5.435 ± 0.042 |
2.443 ± 0.025 | 9.567 ± 0.039 |
2.01 ± 0.019 | 2.893 ± 0.032 |
1.252 ± 0.024 | 10.309 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.678 ± 0.015 | 1.424 ± 0.022 |
6.123 ± 0.035 | 2.531 ± 0.02 |
7.972 ± 0.048 | 4.566 ± 0.028 |
6.084 ± 0.04 | 10.261 ± 0.046 |
1.578 ± 0.019 | 1.776 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
