
Kaistia soli DSM 19436
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Kaistiaceae; Kaistia; Kaistia soli
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
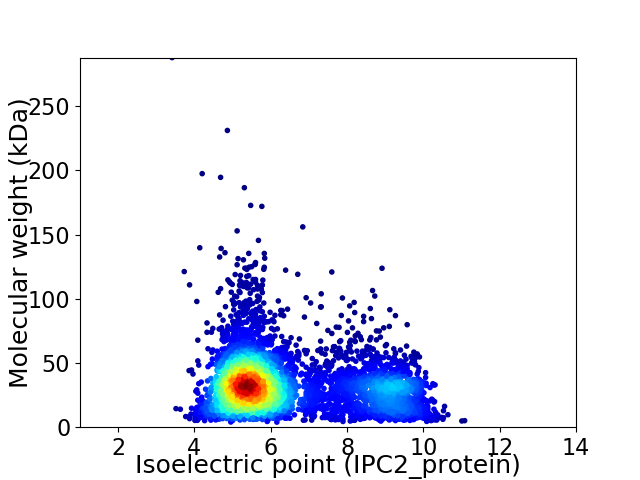
Virtual 2D-PAGE plot for 4866 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
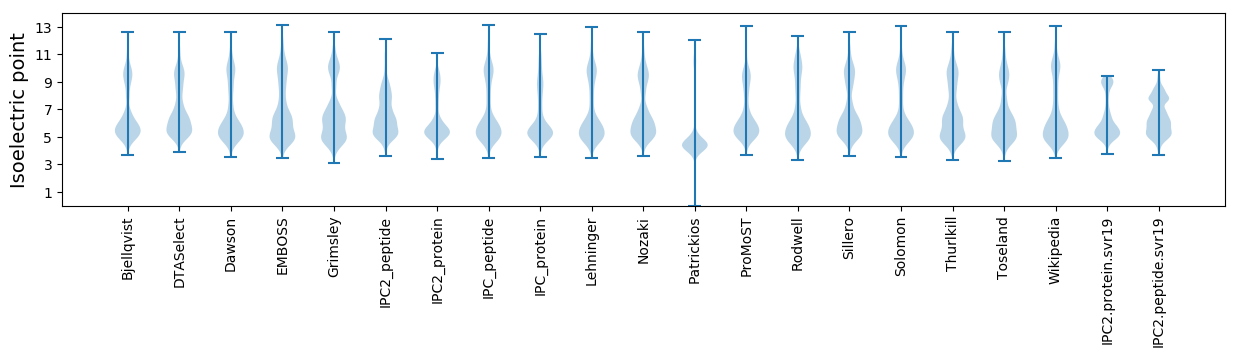
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5IBL2|A0A1M5IBL2_9RHIZ Simple sugar transport system permease protein OS=Kaistia soli DSM 19436 OX=1122133 GN=SAMN02745157_3831 PE=4 SV=1
MM1 pKa = 7.17SQFPASQSGWLPARR15 pKa = 11.84GSALLASASVLALTLGFVPDD35 pKa = 3.74RR36 pKa = 11.84VHH38 pKa = 6.44AAGWNLYY45 pKa = 10.32DD46 pKa = 5.82GYY48 pKa = 8.16TQSYY52 pKa = 8.89SVTYY56 pKa = 10.75ANSLEE61 pKa = 4.1TALADD66 pKa = 4.32KK67 pKa = 9.32DD68 pKa = 4.05TLHH71 pKa = 6.69VGLSVGGSPVTVTMDD86 pKa = 3.2TGSVGLVLSANHH98 pKa = 5.75VASYY102 pKa = 8.59STSGTPGWEE111 pKa = 4.39YY112 pKa = 11.28YY113 pKa = 10.49NSSGLLLTGYY123 pKa = 10.55FNDD126 pKa = 3.8YY127 pKa = 9.19TVEE130 pKa = 4.17LDD132 pKa = 3.82NGTDD136 pKa = 3.43ANGNPTTVTATLPVLVVTEE155 pKa = 4.99AYY157 pKa = 10.06CLGVGSDD164 pKa = 3.59PCDD167 pKa = 3.5AEE169 pKa = 3.97ASKK172 pKa = 11.35NSVSMMGVGYY182 pKa = 10.45DD183 pKa = 3.61RR184 pKa = 11.84NTMGTGSVDD193 pKa = 3.42LSLSGKK199 pKa = 8.08QLNEE203 pKa = 3.93QLNAAPTTSEE213 pKa = 4.44AYY215 pKa = 10.67NLFLNIDD222 pKa = 4.07GMAEE226 pKa = 3.72GALRR230 pKa = 11.84RR231 pKa = 11.84GYY233 pKa = 10.11IITPTGVEE241 pKa = 4.23LGLTAANTSSQAFTYY256 pKa = 10.06AQLVLNSAGTNGATSNWQSVAADD279 pKa = 3.44VTLAGTSSTATLLMDD294 pKa = 3.77TGITGSFFEE303 pKa = 4.79IPGGTEE309 pKa = 3.98GPATAGTVITISLAGGSATYY329 pKa = 10.74SFVVGDD335 pKa = 3.74TANPQTPGTVTIGPPAAAFVNSGLHH360 pKa = 5.64TYY362 pKa = 10.94AGFNVLFDD370 pKa = 4.24ADD372 pKa = 4.02GGFLGVAANGFSGATNASVTQLIAATGPLTLTQAFEE408 pKa = 4.06TDD410 pKa = 4.06LPVMLLDD417 pKa = 4.32ASTINTSTTATFDD430 pKa = 3.17AGIFGPGSLTLNGGTVVLNGAVTNGGGVTAASGTTALNGTMTGNLTVASGASFYY484 pKa = 11.27NYY486 pKa = 10.07NNGYY490 pKa = 10.07AVAAGNILVNDD501 pKa = 4.25GLFVGANSGAAFVNAGTVDD520 pKa = 3.34NSGSFVGAVNNSGSWTNSGTLTGDD544 pKa = 3.35VTNSGTFSNSNLVDD558 pKa = 3.8GNITNTGSLTNTGEE572 pKa = 3.89IEE574 pKa = 4.24GDD576 pKa = 3.7VTSTGPIANQGTVTGTLTVYY596 pKa = 10.1NQHH599 pKa = 6.41SGNGTVGTLSAKK611 pKa = 10.19PGALVSPGNSVGTIIVSGDD630 pKa = 2.98ATFEE634 pKa = 4.55PGSVLYY640 pKa = 10.88AEE642 pKa = 5.53LGANGLSDD650 pKa = 4.17LLVVGGTLVADD661 pKa = 4.67GATLYY666 pKa = 10.68LAAANGFEE674 pKa = 4.15PVLGNSYY681 pKa = 11.14SVIQAGSIASNFTVASPFFGSTASPFPFLGASLDD715 pKa = 3.82GTGVLTLGRR724 pKa = 11.84SALRR728 pKa = 11.84FEE730 pKa = 5.26DD731 pKa = 4.6FAVTQNEE738 pKa = 3.89RR739 pKa = 11.84MAASAAEE746 pKa = 4.1TLGLQSPLNQALALMSIAEE765 pKa = 4.3VPSVFDD771 pKa = 3.66SLSGEE776 pKa = 3.89IAASAEE782 pKa = 4.23STLQQQSIYY791 pKa = 10.71LRR793 pKa = 11.84DD794 pKa = 3.47AVTGRR799 pKa = 11.84VRR801 pKa = 11.84QAFSDD806 pKa = 3.62AAGPEE811 pKa = 4.08ASGSQTARR819 pKa = 11.84LAPGLDD825 pKa = 3.08ATAWTQAYY833 pKa = 7.81GAWGNSWSDD842 pKa = 3.2GNAAAVSRR850 pKa = 11.84SIGGFLLGADD860 pKa = 4.11AALGDD865 pKa = 3.75AWRR868 pKa = 11.84VGLAGGYY875 pKa = 8.16SQSDD879 pKa = 3.66FSLDD883 pKa = 3.42GVGGGGTSDD892 pKa = 3.67NYY894 pKa = 11.34DD895 pKa = 3.18VAIYY899 pKa = 10.46GGTRR903 pKa = 11.84QGDD906 pKa = 3.04ASLRR910 pKa = 11.84FGAGYY915 pKa = 7.69TWHH918 pKa = 7.88DD919 pKa = 3.27IATGRR924 pKa = 11.84TALLPTTAEE933 pKa = 4.26FLSADD938 pKa = 3.95YY939 pKa = 10.89QGGTAQVFGEE949 pKa = 3.96AAYY952 pKa = 10.14DD953 pKa = 3.63VRR955 pKa = 11.84LGRR958 pKa = 11.84AVLEE962 pKa = 4.57PYY964 pKa = 10.99VNLAYY969 pKa = 11.05VNLTMDD975 pKa = 4.65GFWEE979 pKa = 4.33TGGAAALTFAEE990 pKa = 4.67STMSTTFNVLGMRR1003 pKa = 11.84LGQAFDD1009 pKa = 3.67IGNGLQLLTRR1019 pKa = 11.84GSLGWQHH1026 pKa = 6.8AFGDD1030 pKa = 3.88ITPQATAAFLGSSAFTVAGLPIAQDD1055 pKa = 3.19AALIDD1060 pKa = 3.72AFIGFRR1066 pKa = 11.84PTSRR1070 pKa = 11.84VDD1072 pKa = 3.57FGLRR1076 pKa = 11.84YY1077 pKa = 9.89SGQIADD1083 pKa = 4.64DD1084 pKa = 3.82ATDD1087 pKa = 3.46NAVQGTLDD1095 pKa = 3.11IRR1097 pKa = 11.84FF1098 pKa = 3.63
MM1 pKa = 7.17SQFPASQSGWLPARR15 pKa = 11.84GSALLASASVLALTLGFVPDD35 pKa = 3.74RR36 pKa = 11.84VHH38 pKa = 6.44AAGWNLYY45 pKa = 10.32DD46 pKa = 5.82GYY48 pKa = 8.16TQSYY52 pKa = 8.89SVTYY56 pKa = 10.75ANSLEE61 pKa = 4.1TALADD66 pKa = 4.32KK67 pKa = 9.32DD68 pKa = 4.05TLHH71 pKa = 6.69VGLSVGGSPVTVTMDD86 pKa = 3.2TGSVGLVLSANHH98 pKa = 5.75VASYY102 pKa = 8.59STSGTPGWEE111 pKa = 4.39YY112 pKa = 11.28YY113 pKa = 10.49NSSGLLLTGYY123 pKa = 10.55FNDD126 pKa = 3.8YY127 pKa = 9.19TVEE130 pKa = 4.17LDD132 pKa = 3.82NGTDD136 pKa = 3.43ANGNPTTVTATLPVLVVTEE155 pKa = 4.99AYY157 pKa = 10.06CLGVGSDD164 pKa = 3.59PCDD167 pKa = 3.5AEE169 pKa = 3.97ASKK172 pKa = 11.35NSVSMMGVGYY182 pKa = 10.45DD183 pKa = 3.61RR184 pKa = 11.84NTMGTGSVDD193 pKa = 3.42LSLSGKK199 pKa = 8.08QLNEE203 pKa = 3.93QLNAAPTTSEE213 pKa = 4.44AYY215 pKa = 10.67NLFLNIDD222 pKa = 4.07GMAEE226 pKa = 3.72GALRR230 pKa = 11.84RR231 pKa = 11.84GYY233 pKa = 10.11IITPTGVEE241 pKa = 4.23LGLTAANTSSQAFTYY256 pKa = 10.06AQLVLNSAGTNGATSNWQSVAADD279 pKa = 3.44VTLAGTSSTATLLMDD294 pKa = 3.77TGITGSFFEE303 pKa = 4.79IPGGTEE309 pKa = 3.98GPATAGTVITISLAGGSATYY329 pKa = 10.74SFVVGDD335 pKa = 3.74TANPQTPGTVTIGPPAAAFVNSGLHH360 pKa = 5.64TYY362 pKa = 10.94AGFNVLFDD370 pKa = 4.24ADD372 pKa = 4.02GGFLGVAANGFSGATNASVTQLIAATGPLTLTQAFEE408 pKa = 4.06TDD410 pKa = 4.06LPVMLLDD417 pKa = 4.32ASTINTSTTATFDD430 pKa = 3.17AGIFGPGSLTLNGGTVVLNGAVTNGGGVTAASGTTALNGTMTGNLTVASGASFYY484 pKa = 11.27NYY486 pKa = 10.07NNGYY490 pKa = 10.07AVAAGNILVNDD501 pKa = 4.25GLFVGANSGAAFVNAGTVDD520 pKa = 3.34NSGSFVGAVNNSGSWTNSGTLTGDD544 pKa = 3.35VTNSGTFSNSNLVDD558 pKa = 3.8GNITNTGSLTNTGEE572 pKa = 3.89IEE574 pKa = 4.24GDD576 pKa = 3.7VTSTGPIANQGTVTGTLTVYY596 pKa = 10.1NQHH599 pKa = 6.41SGNGTVGTLSAKK611 pKa = 10.19PGALVSPGNSVGTIIVSGDD630 pKa = 2.98ATFEE634 pKa = 4.55PGSVLYY640 pKa = 10.88AEE642 pKa = 5.53LGANGLSDD650 pKa = 4.17LLVVGGTLVADD661 pKa = 4.67GATLYY666 pKa = 10.68LAAANGFEE674 pKa = 4.15PVLGNSYY681 pKa = 11.14SVIQAGSIASNFTVASPFFGSTASPFPFLGASLDD715 pKa = 3.82GTGVLTLGRR724 pKa = 11.84SALRR728 pKa = 11.84FEE730 pKa = 5.26DD731 pKa = 4.6FAVTQNEE738 pKa = 3.89RR739 pKa = 11.84MAASAAEE746 pKa = 4.1TLGLQSPLNQALALMSIAEE765 pKa = 4.3VPSVFDD771 pKa = 3.66SLSGEE776 pKa = 3.89IAASAEE782 pKa = 4.23STLQQQSIYY791 pKa = 10.71LRR793 pKa = 11.84DD794 pKa = 3.47AVTGRR799 pKa = 11.84VRR801 pKa = 11.84QAFSDD806 pKa = 3.62AAGPEE811 pKa = 4.08ASGSQTARR819 pKa = 11.84LAPGLDD825 pKa = 3.08ATAWTQAYY833 pKa = 7.81GAWGNSWSDD842 pKa = 3.2GNAAAVSRR850 pKa = 11.84SIGGFLLGADD860 pKa = 4.11AALGDD865 pKa = 3.75AWRR868 pKa = 11.84VGLAGGYY875 pKa = 8.16SQSDD879 pKa = 3.66FSLDD883 pKa = 3.42GVGGGGTSDD892 pKa = 3.67NYY894 pKa = 11.34DD895 pKa = 3.18VAIYY899 pKa = 10.46GGTRR903 pKa = 11.84QGDD906 pKa = 3.04ASLRR910 pKa = 11.84FGAGYY915 pKa = 7.69TWHH918 pKa = 7.88DD919 pKa = 3.27IATGRR924 pKa = 11.84TALLPTTAEE933 pKa = 4.26FLSADD938 pKa = 3.95YY939 pKa = 10.89QGGTAQVFGEE949 pKa = 3.96AAYY952 pKa = 10.14DD953 pKa = 3.63VRR955 pKa = 11.84LGRR958 pKa = 11.84AVLEE962 pKa = 4.57PYY964 pKa = 10.99VNLAYY969 pKa = 11.05VNLTMDD975 pKa = 4.65GFWEE979 pKa = 4.33TGGAAALTFAEE990 pKa = 4.67STMSTTFNVLGMRR1003 pKa = 11.84LGQAFDD1009 pKa = 3.67IGNGLQLLTRR1019 pKa = 11.84GSLGWQHH1026 pKa = 6.8AFGDD1030 pKa = 3.88ITPQATAAFLGSSAFTVAGLPIAQDD1055 pKa = 3.19AALIDD1060 pKa = 3.72AFIGFRR1066 pKa = 11.84PTSRR1070 pKa = 11.84VDD1072 pKa = 3.57FGLRR1076 pKa = 11.84YY1077 pKa = 9.89SGQIADD1083 pKa = 4.64DD1084 pKa = 3.82ATDD1087 pKa = 3.46NAVQGTLDD1095 pKa = 3.11IRR1097 pKa = 11.84FF1098 pKa = 3.63
Molecular weight: 110.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5NXS0|A0A1M5NXS0_9RHIZ Uncharacterized protein OS=Kaistia soli DSM 19436 OX=1122133 GN=SAMN02745157_0039 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.36VIAQRR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.47KK41 pKa = 9.66LTAA44 pKa = 4.17
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.36VIAQRR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.47KK41 pKa = 9.66LTAA44 pKa = 4.17
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1558496 |
35 |
3023 |
320.3 |
34.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.309 ± 0.05 | 0.69 ± 0.011 |
5.77 ± 0.026 | 5.326 ± 0.034 |
3.821 ± 0.025 | 8.841 ± 0.042 |
1.951 ± 0.016 | 5.536 ± 0.022 |
2.898 ± 0.026 | 10.126 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.356 ± 0.015 | 2.398 ± 0.023 |
5.379 ± 0.027 | 2.722 ± 0.018 |
7.021 ± 0.04 | 5.454 ± 0.027 |
5.349 ± 0.027 | 7.595 ± 0.029 |
1.319 ± 0.015 | 2.137 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
