
Citrus yellow vein-associated virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.91
Get precalculated fractions of proteins
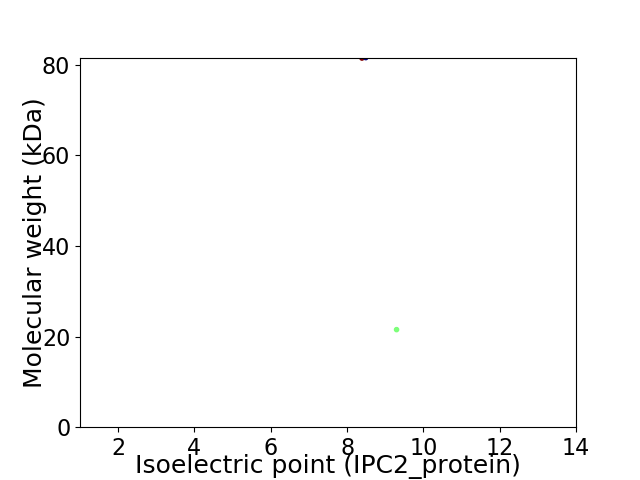
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S4TYN1|S4TYN1_9VIRU RNA-dependent RNA polymerase OS=Citrus yellow vein-associated virus OX=1297894 GN=RdRp PE=4 SV=1
MM1 pKa = 7.98DD2 pKa = 5.76PSSLPRR8 pKa = 11.84ACWHH12 pKa = 6.7HH13 pKa = 5.69ARR15 pKa = 11.84QVFRR19 pKa = 11.84ASTSFSRR26 pKa = 11.84EE27 pKa = 3.47VVRR30 pKa = 11.84AARR33 pKa = 11.84RR34 pKa = 11.84WVTMPTSRR42 pKa = 11.84HH43 pKa = 5.9AYY45 pKa = 10.06DD46 pKa = 4.59FSTSLGIVIAEE57 pKa = 3.76PAARR61 pKa = 11.84LRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84LPSVRR70 pKa = 11.84KK71 pKa = 8.98CAEE74 pKa = 3.73KK75 pKa = 10.7LVVHH79 pKa = 6.16KK80 pKa = 10.82QVDD83 pKa = 3.81TLVDD87 pKa = 3.92EE88 pKa = 4.65WCSGIPNPDD97 pKa = 2.89IVEE100 pKa = 4.32VGWALRR106 pKa = 11.84LRR108 pKa = 11.84DD109 pKa = 3.99RR110 pKa = 11.84FGLPPASEE118 pKa = 4.22PTRR121 pKa = 11.84LSGEE125 pKa = 3.92RR126 pKa = 11.84WVLKK130 pKa = 9.92QLNGVDD136 pKa = 3.9PEE138 pKa = 4.35SWNADD143 pKa = 3.26LGRR146 pKa = 11.84SVHH149 pKa = 5.76IQGDD153 pKa = 3.93YY154 pKa = 10.82APGRR158 pKa = 11.84NAHH161 pKa = 6.39IAQVAATLWLTRR173 pKa = 11.84TLHH176 pKa = 6.64DD177 pKa = 3.77KK178 pKa = 10.98ALARR182 pKa = 11.84HH183 pKa = 5.48QGFSRR188 pKa = 11.84FAVIGVDD195 pKa = 3.42GLEE198 pKa = 4.24AKK200 pKa = 10.01AVPLASGLRR209 pKa = 11.84LLPVPRR215 pKa = 11.84PGQSRR220 pKa = 11.84RR221 pKa = 11.84LSQTLLLPTPRR232 pKa = 11.84AQFVNHH238 pKa = 6.64ACSANNLGRR247 pKa = 11.84VMTTRR252 pKa = 11.84VLQYY256 pKa = 10.5KK257 pKa = 10.18GRR259 pKa = 11.84DD260 pKa = 3.56PILPSSEE267 pKa = 3.61ALHH270 pKa = 6.2RR271 pKa = 11.84LNLRR275 pKa = 11.84IAEE278 pKa = 4.81LYY280 pKa = 9.94RR281 pKa = 11.84SRR283 pKa = 11.84PSTVYY288 pKa = 9.2PLSYY292 pKa = 10.7EE293 pKa = 4.16GFLNCYY299 pKa = 8.89EE300 pKa = 3.94GRR302 pKa = 11.84QRR304 pKa = 11.84TRR306 pKa = 11.84YY307 pKa = 8.48AQAVEE312 pKa = 3.68QLMRR316 pKa = 11.84STLEE320 pKa = 4.01PKK322 pKa = 10.12DD323 pKa = 3.55ARR325 pKa = 11.84VEE327 pKa = 4.07TFIKK331 pKa = 10.67NEE333 pKa = 3.87KK334 pKa = 9.72FDD336 pKa = 3.53WALKK340 pKa = 10.68GEE342 pKa = 4.35EE343 pKa = 4.24ADD345 pKa = 3.84PRR347 pKa = 11.84AIQPRR352 pKa = 11.84KK353 pKa = 8.79PKK355 pKa = 10.37YY356 pKa = 8.89LAEE359 pKa = 4.02VGRR362 pKa = 11.84WFKK365 pKa = 10.32PLEE368 pKa = 4.24RR369 pKa = 11.84IIYY372 pKa = 9.51KK373 pKa = 10.36DD374 pKa = 3.27LSKK377 pKa = 10.93RR378 pKa = 11.84LYY380 pKa = 10.97GEE382 pKa = 4.06GAEE385 pKa = 4.01PCIAKK390 pKa = 10.12GLNALEE396 pKa = 4.46SGATLRR402 pKa = 11.84RR403 pKa = 11.84KK404 pKa = 8.47WEE406 pKa = 4.06KK407 pKa = 10.23FSSPVCVSLDD417 pKa = 3.2ASRR420 pKa = 11.84FDD422 pKa = 3.5LHH424 pKa = 8.4VSVGMLKK431 pKa = 9.18FTHH434 pKa = 6.73KK435 pKa = 10.79LYY437 pKa = 10.56DD438 pKa = 4.02YY439 pKa = 9.48YY440 pKa = 11.19CKK442 pKa = 10.69SPTLQRR448 pKa = 11.84YY449 pKa = 7.53LKK451 pKa = 8.41WTLRR455 pKa = 11.84NHH457 pKa = 6.55GVASCKK463 pKa = 9.82EE464 pKa = 3.64LSYY467 pKa = 10.5EE468 pKa = 4.07YY469 pKa = 10.27EE470 pKa = 4.18VVGRR474 pKa = 11.84RR475 pKa = 11.84MSGDD479 pKa = 3.17MDD481 pKa = 3.91TALGNCVIMSILTWFMLSEE500 pKa = 4.28LGIKK504 pKa = 10.13HH505 pKa = 6.2EE506 pKa = 4.74LFDD509 pKa = 5.42NGDD512 pKa = 3.56DD513 pKa = 4.05CLFICEE519 pKa = 4.39SHH521 pKa = 7.08DD522 pKa = 3.65VPSPEE527 pKa = 5.54VITNWFSDD535 pKa = 3.25FGFVVRR541 pKa = 11.84LEE543 pKa = 4.33GVTSVFEE550 pKa = 4.61RR551 pKa = 11.84IEE553 pKa = 4.15FCQTSPVWTEE563 pKa = 4.19RR564 pKa = 11.84GWLMCRR570 pKa = 11.84NIKK573 pKa = 10.27SLSKK577 pKa = 10.89DD578 pKa = 3.19LTNVNSCTGSTIEE591 pKa = 3.89YY592 pKa = 6.55THH594 pKa = 6.68WLKK597 pKa = 11.0AVGKK601 pKa = 9.92CGSILNAGVPIFQSFHH617 pKa = 6.42NMLEE621 pKa = 4.05RR622 pKa = 11.84LGTNSRR628 pKa = 11.84IDD630 pKa = 2.92RR631 pKa = 11.84GVFFKK636 pKa = 11.0SGLVNLIRR644 pKa = 11.84GMDD647 pKa = 3.63RR648 pKa = 11.84QPDD651 pKa = 3.44VDD653 pKa = 3.35ITTSARR659 pKa = 11.84LSFEE663 pKa = 4.05VAFGITPGMQLAIEE677 pKa = 4.82RR678 pKa = 11.84YY679 pKa = 8.31YY680 pKa = 11.29DD681 pKa = 3.5SVMGSLSKK689 pKa = 10.56IEE691 pKa = 4.06TTKK694 pKa = 10.43WPIEE698 pKa = 3.71LRR700 pKa = 11.84KK701 pKa = 9.8EE702 pKa = 4.1YY703 pKa = 10.54EE704 pKa = 4.77HH705 pKa = 7.63GSEE708 pKa = 3.94WYY710 pKa = 9.73EE711 pKa = 3.81DD712 pKa = 3.71LGVLGG717 pKa = 4.78
MM1 pKa = 7.98DD2 pKa = 5.76PSSLPRR8 pKa = 11.84ACWHH12 pKa = 6.7HH13 pKa = 5.69ARR15 pKa = 11.84QVFRR19 pKa = 11.84ASTSFSRR26 pKa = 11.84EE27 pKa = 3.47VVRR30 pKa = 11.84AARR33 pKa = 11.84RR34 pKa = 11.84WVTMPTSRR42 pKa = 11.84HH43 pKa = 5.9AYY45 pKa = 10.06DD46 pKa = 4.59FSTSLGIVIAEE57 pKa = 3.76PAARR61 pKa = 11.84LRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84LPSVRR70 pKa = 11.84KK71 pKa = 8.98CAEE74 pKa = 3.73KK75 pKa = 10.7LVVHH79 pKa = 6.16KK80 pKa = 10.82QVDD83 pKa = 3.81TLVDD87 pKa = 3.92EE88 pKa = 4.65WCSGIPNPDD97 pKa = 2.89IVEE100 pKa = 4.32VGWALRR106 pKa = 11.84LRR108 pKa = 11.84DD109 pKa = 3.99RR110 pKa = 11.84FGLPPASEE118 pKa = 4.22PTRR121 pKa = 11.84LSGEE125 pKa = 3.92RR126 pKa = 11.84WVLKK130 pKa = 9.92QLNGVDD136 pKa = 3.9PEE138 pKa = 4.35SWNADD143 pKa = 3.26LGRR146 pKa = 11.84SVHH149 pKa = 5.76IQGDD153 pKa = 3.93YY154 pKa = 10.82APGRR158 pKa = 11.84NAHH161 pKa = 6.39IAQVAATLWLTRR173 pKa = 11.84TLHH176 pKa = 6.64DD177 pKa = 3.77KK178 pKa = 10.98ALARR182 pKa = 11.84HH183 pKa = 5.48QGFSRR188 pKa = 11.84FAVIGVDD195 pKa = 3.42GLEE198 pKa = 4.24AKK200 pKa = 10.01AVPLASGLRR209 pKa = 11.84LLPVPRR215 pKa = 11.84PGQSRR220 pKa = 11.84RR221 pKa = 11.84LSQTLLLPTPRR232 pKa = 11.84AQFVNHH238 pKa = 6.64ACSANNLGRR247 pKa = 11.84VMTTRR252 pKa = 11.84VLQYY256 pKa = 10.5KK257 pKa = 10.18GRR259 pKa = 11.84DD260 pKa = 3.56PILPSSEE267 pKa = 3.61ALHH270 pKa = 6.2RR271 pKa = 11.84LNLRR275 pKa = 11.84IAEE278 pKa = 4.81LYY280 pKa = 9.94RR281 pKa = 11.84SRR283 pKa = 11.84PSTVYY288 pKa = 9.2PLSYY292 pKa = 10.7EE293 pKa = 4.16GFLNCYY299 pKa = 8.89EE300 pKa = 3.94GRR302 pKa = 11.84QRR304 pKa = 11.84TRR306 pKa = 11.84YY307 pKa = 8.48AQAVEE312 pKa = 3.68QLMRR316 pKa = 11.84STLEE320 pKa = 4.01PKK322 pKa = 10.12DD323 pKa = 3.55ARR325 pKa = 11.84VEE327 pKa = 4.07TFIKK331 pKa = 10.67NEE333 pKa = 3.87KK334 pKa = 9.72FDD336 pKa = 3.53WALKK340 pKa = 10.68GEE342 pKa = 4.35EE343 pKa = 4.24ADD345 pKa = 3.84PRR347 pKa = 11.84AIQPRR352 pKa = 11.84KK353 pKa = 8.79PKK355 pKa = 10.37YY356 pKa = 8.89LAEE359 pKa = 4.02VGRR362 pKa = 11.84WFKK365 pKa = 10.32PLEE368 pKa = 4.24RR369 pKa = 11.84IIYY372 pKa = 9.51KK373 pKa = 10.36DD374 pKa = 3.27LSKK377 pKa = 10.93RR378 pKa = 11.84LYY380 pKa = 10.97GEE382 pKa = 4.06GAEE385 pKa = 4.01PCIAKK390 pKa = 10.12GLNALEE396 pKa = 4.46SGATLRR402 pKa = 11.84RR403 pKa = 11.84KK404 pKa = 8.47WEE406 pKa = 4.06KK407 pKa = 10.23FSSPVCVSLDD417 pKa = 3.2ASRR420 pKa = 11.84FDD422 pKa = 3.5LHH424 pKa = 8.4VSVGMLKK431 pKa = 9.18FTHH434 pKa = 6.73KK435 pKa = 10.79LYY437 pKa = 10.56DD438 pKa = 4.02YY439 pKa = 9.48YY440 pKa = 11.19CKK442 pKa = 10.69SPTLQRR448 pKa = 11.84YY449 pKa = 7.53LKK451 pKa = 8.41WTLRR455 pKa = 11.84NHH457 pKa = 6.55GVASCKK463 pKa = 9.82EE464 pKa = 3.64LSYY467 pKa = 10.5EE468 pKa = 4.07YY469 pKa = 10.27EE470 pKa = 4.18VVGRR474 pKa = 11.84RR475 pKa = 11.84MSGDD479 pKa = 3.17MDD481 pKa = 3.91TALGNCVIMSILTWFMLSEE500 pKa = 4.28LGIKK504 pKa = 10.13HH505 pKa = 6.2EE506 pKa = 4.74LFDD509 pKa = 5.42NGDD512 pKa = 3.56DD513 pKa = 4.05CLFICEE519 pKa = 4.39SHH521 pKa = 7.08DD522 pKa = 3.65VPSPEE527 pKa = 5.54VITNWFSDD535 pKa = 3.25FGFVVRR541 pKa = 11.84LEE543 pKa = 4.33GVTSVFEE550 pKa = 4.61RR551 pKa = 11.84IEE553 pKa = 4.15FCQTSPVWTEE563 pKa = 4.19RR564 pKa = 11.84GWLMCRR570 pKa = 11.84NIKK573 pKa = 10.27SLSKK577 pKa = 10.89DD578 pKa = 3.19LTNVNSCTGSTIEE591 pKa = 3.89YY592 pKa = 6.55THH594 pKa = 6.68WLKK597 pKa = 11.0AVGKK601 pKa = 9.92CGSILNAGVPIFQSFHH617 pKa = 6.42NMLEE621 pKa = 4.05RR622 pKa = 11.84LGTNSRR628 pKa = 11.84IDD630 pKa = 2.92RR631 pKa = 11.84GVFFKK636 pKa = 11.0SGLVNLIRR644 pKa = 11.84GMDD647 pKa = 3.63RR648 pKa = 11.84QPDD651 pKa = 3.44VDD653 pKa = 3.35ITTSARR659 pKa = 11.84LSFEE663 pKa = 4.05VAFGITPGMQLAIEE677 pKa = 4.82RR678 pKa = 11.84YY679 pKa = 8.31YY680 pKa = 11.29DD681 pKa = 3.5SVMGSLSKK689 pKa = 10.56IEE691 pKa = 4.06TTKK694 pKa = 10.43WPIEE698 pKa = 3.71LRR700 pKa = 11.84KK701 pKa = 9.8EE702 pKa = 4.1YY703 pKa = 10.54EE704 pKa = 4.77HH705 pKa = 7.63GSEE708 pKa = 3.94WYY710 pKa = 9.73EE711 pKa = 3.81DD712 pKa = 3.71LGVLGG717 pKa = 4.78
Molecular weight: 81.44 kDa
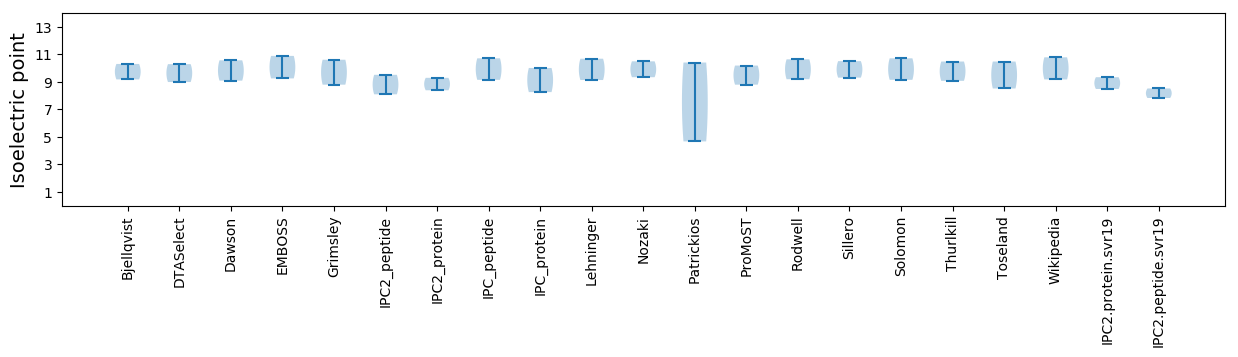
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S4TYN1|S4TYN1_9VIRU RNA-dependent RNA polymerase OS=Citrus yellow vein-associated virus OX=1297894 GN=RdRp PE=4 SV=1
MM1 pKa = 7.98DD2 pKa = 5.76PSSLPRR8 pKa = 11.84ACWHH12 pKa = 6.7HH13 pKa = 5.69ARR15 pKa = 11.84QVFRR19 pKa = 11.84ASTSFSRR26 pKa = 11.84EE27 pKa = 3.47VVRR30 pKa = 11.84AARR33 pKa = 11.84RR34 pKa = 11.84WVTMPTSRR42 pKa = 11.84HH43 pKa = 5.9AYY45 pKa = 10.06DD46 pKa = 4.59FSTSLGIVIAEE57 pKa = 3.76PAARR61 pKa = 11.84LRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84LPSVRR70 pKa = 11.84KK71 pKa = 8.98CAEE74 pKa = 3.73KK75 pKa = 10.7LVVHH79 pKa = 6.16KK80 pKa = 10.82QVDD83 pKa = 3.81TLVDD87 pKa = 3.92EE88 pKa = 4.65WCSGIPNPDD97 pKa = 2.89IVEE100 pKa = 4.32VGWALRR106 pKa = 11.84LRR108 pKa = 11.84DD109 pKa = 3.99RR110 pKa = 11.84FGLPPASEE118 pKa = 4.22PTRR121 pKa = 11.84LSGEE125 pKa = 3.92RR126 pKa = 11.84WVLKK130 pKa = 9.92QLNGVDD136 pKa = 3.9PEE138 pKa = 4.35SWNADD143 pKa = 3.26LGRR146 pKa = 11.84SVHH149 pKa = 5.76IQGDD153 pKa = 3.93YY154 pKa = 10.82APGRR158 pKa = 11.84NAHH161 pKa = 6.39IAQVAATLWLTRR173 pKa = 11.84TLHH176 pKa = 6.64DD177 pKa = 3.77KK178 pKa = 10.98ALARR182 pKa = 11.84HH183 pKa = 5.38QGFRR187 pKa = 11.84DD188 pKa = 3.62LQQ190 pKa = 3.74
MM1 pKa = 7.98DD2 pKa = 5.76PSSLPRR8 pKa = 11.84ACWHH12 pKa = 6.7HH13 pKa = 5.69ARR15 pKa = 11.84QVFRR19 pKa = 11.84ASTSFSRR26 pKa = 11.84EE27 pKa = 3.47VVRR30 pKa = 11.84AARR33 pKa = 11.84RR34 pKa = 11.84WVTMPTSRR42 pKa = 11.84HH43 pKa = 5.9AYY45 pKa = 10.06DD46 pKa = 4.59FSTSLGIVIAEE57 pKa = 3.76PAARR61 pKa = 11.84LRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84LPSVRR70 pKa = 11.84KK71 pKa = 8.98CAEE74 pKa = 3.73KK75 pKa = 10.7LVVHH79 pKa = 6.16KK80 pKa = 10.82QVDD83 pKa = 3.81TLVDD87 pKa = 3.92EE88 pKa = 4.65WCSGIPNPDD97 pKa = 2.89IVEE100 pKa = 4.32VGWALRR106 pKa = 11.84LRR108 pKa = 11.84DD109 pKa = 3.99RR110 pKa = 11.84FGLPPASEE118 pKa = 4.22PTRR121 pKa = 11.84LSGEE125 pKa = 3.92RR126 pKa = 11.84WVLKK130 pKa = 9.92QLNGVDD136 pKa = 3.9PEE138 pKa = 4.35SWNADD143 pKa = 3.26LGRR146 pKa = 11.84SVHH149 pKa = 5.76IQGDD153 pKa = 3.93YY154 pKa = 10.82APGRR158 pKa = 11.84NAHH161 pKa = 6.39IAQVAATLWLTRR173 pKa = 11.84TLHH176 pKa = 6.64DD177 pKa = 3.77KK178 pKa = 10.98ALARR182 pKa = 11.84HH183 pKa = 5.38QGFRR187 pKa = 11.84DD188 pKa = 3.62LQQ190 pKa = 3.74
Molecular weight: 21.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
907 |
190 |
717 |
453.5 |
51.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.497 ± 1.509 | 2.095 ± 0.257 |
4.851 ± 0.467 | 5.954 ± 0.868 |
3.638 ± 0.502 | 6.395 ± 0.564 |
2.867 ± 0.669 | 4.079 ± 0.459 |
4.19 ± 0.776 | 10.254 ± 0.389 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.764 ± 0.354 | 2.867 ± 0.379 |
5.402 ± 0.455 | 2.867 ± 0.407 |
9.592 ± 1.252 | 7.828 ± 0.229 |
5.072 ± 0.167 | 7.387 ± 0.515 |
2.756 ± 0.462 | 2.646 ± 0.794 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
