
Opitutaceae bacterium TSB47
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Opitutae; Opitutales; Opitutaceae; unclassified Opitutaceae
Average proteome isoelectric point is 7.12
Get precalculated fractions of proteins
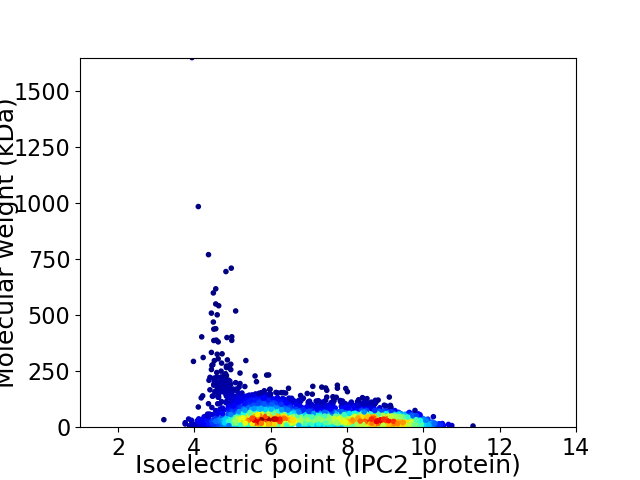
Virtual 2D-PAGE plot for 4993 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
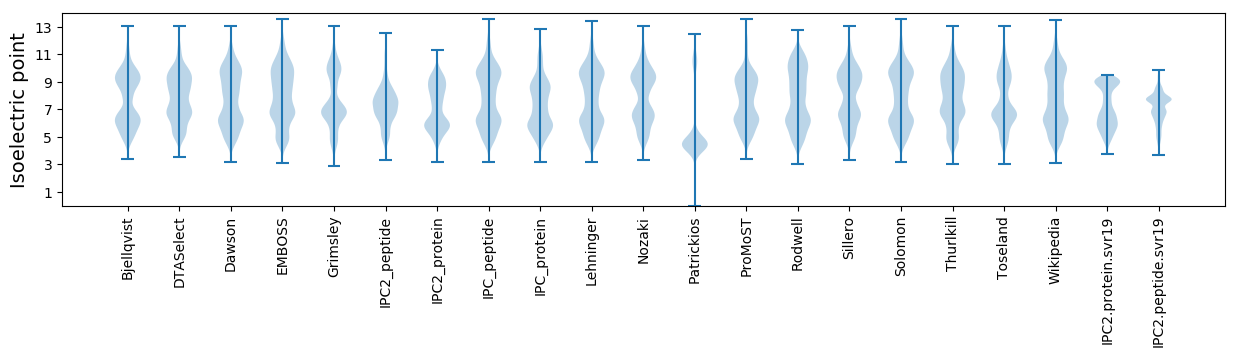
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A178IN72|A0A178IN72_9BACT Beta-N-acetylhexosaminidase OS=Opitutaceae bacterium TSB47 OX=1184151 GN=AW736_07695 PE=3 SV=1
MM1 pKa = 7.42IKK3 pKa = 10.4NVFKK7 pKa = 11.09LGGVAISALLLSAPAFADD25 pKa = 3.36IPLHH29 pKa = 6.18QNFSANGYY37 pKa = 8.7VAGSYY42 pKa = 7.79MWTDD46 pKa = 3.51KK47 pKa = 11.24GDD49 pKa = 3.62SDD51 pKa = 5.83HH52 pKa = 7.28YY53 pKa = 11.52DD54 pKa = 3.43LDD56 pKa = 3.89VAKK59 pKa = 10.7LGFSINYY66 pKa = 9.21APTKK70 pKa = 10.47AYY72 pKa = 9.73ISLFYY77 pKa = 11.02DD78 pKa = 3.47GDD80 pKa = 4.1EE81 pKa = 4.32INVLDD86 pKa = 5.22AYY88 pKa = 11.18VDD90 pKa = 3.67YY91 pKa = 11.35DD92 pKa = 3.96FGNGAVVTGGRR103 pKa = 11.84FLSFLGFEE111 pKa = 4.78AFDD114 pKa = 4.24APNMYY119 pKa = 10.17QISYY123 pKa = 11.16ANGDD127 pKa = 3.89LFGGSALEE135 pKa = 3.98IAPIPAYY142 pKa = 10.17HH143 pKa = 6.45SGVKK147 pKa = 9.62VVYY150 pKa = 10.73SDD152 pKa = 4.48DD153 pKa = 3.24AWSAGVAFLDD163 pKa = 4.18SVYY166 pKa = 11.11GGLKK170 pKa = 10.35GDD172 pKa = 4.99GEE174 pKa = 4.54VEE176 pKa = 4.32DD177 pKa = 4.19NFGVEE182 pKa = 4.31AFFSFTGVEE191 pKa = 4.01KK192 pKa = 10.1LTLFGGIAFQSEE204 pKa = 4.3DD205 pKa = 4.2DD206 pKa = 4.91DD207 pKa = 4.17PTDD210 pKa = 4.35GYY212 pKa = 9.6TSPEE216 pKa = 3.28IMTFNFWASYY226 pKa = 9.65EE227 pKa = 4.29ISDD230 pKa = 3.35KK231 pKa = 10.81WLVAGEE237 pKa = 4.4FTFSDD242 pKa = 3.29QKK244 pKa = 11.74DD245 pKa = 3.8YY246 pKa = 11.14MDD248 pKa = 4.66GYY250 pKa = 11.4NWLALAKK257 pKa = 10.37YY258 pKa = 10.02AITPKK263 pKa = 9.6ISAVFRR269 pKa = 11.84VSGEE273 pKa = 3.96DD274 pKa = 3.12VDD276 pKa = 4.8YY277 pKa = 10.92EE278 pKa = 4.59DD279 pKa = 6.15PIEE282 pKa = 4.29TDD284 pKa = 3.14PSFLKK289 pKa = 9.88FTLAPAYY296 pKa = 8.68TITDD300 pKa = 3.79NLSVTAEE307 pKa = 3.84VSYY310 pKa = 11.43YY311 pKa = 10.54DD312 pKa = 3.59YY313 pKa = 11.33KK314 pKa = 11.48DD315 pKa = 3.69YY316 pKa = 11.31VVKK319 pKa = 10.76DD320 pKa = 3.69DD321 pKa = 4.05VFVGVQAIFKK331 pKa = 9.48FF332 pKa = 3.98
MM1 pKa = 7.42IKK3 pKa = 10.4NVFKK7 pKa = 11.09LGGVAISALLLSAPAFADD25 pKa = 3.36IPLHH29 pKa = 6.18QNFSANGYY37 pKa = 8.7VAGSYY42 pKa = 7.79MWTDD46 pKa = 3.51KK47 pKa = 11.24GDD49 pKa = 3.62SDD51 pKa = 5.83HH52 pKa = 7.28YY53 pKa = 11.52DD54 pKa = 3.43LDD56 pKa = 3.89VAKK59 pKa = 10.7LGFSINYY66 pKa = 9.21APTKK70 pKa = 10.47AYY72 pKa = 9.73ISLFYY77 pKa = 11.02DD78 pKa = 3.47GDD80 pKa = 4.1EE81 pKa = 4.32INVLDD86 pKa = 5.22AYY88 pKa = 11.18VDD90 pKa = 3.67YY91 pKa = 11.35DD92 pKa = 3.96FGNGAVVTGGRR103 pKa = 11.84FLSFLGFEE111 pKa = 4.78AFDD114 pKa = 4.24APNMYY119 pKa = 10.17QISYY123 pKa = 11.16ANGDD127 pKa = 3.89LFGGSALEE135 pKa = 3.98IAPIPAYY142 pKa = 10.17HH143 pKa = 6.45SGVKK147 pKa = 9.62VVYY150 pKa = 10.73SDD152 pKa = 4.48DD153 pKa = 3.24AWSAGVAFLDD163 pKa = 4.18SVYY166 pKa = 11.11GGLKK170 pKa = 10.35GDD172 pKa = 4.99GEE174 pKa = 4.54VEE176 pKa = 4.32DD177 pKa = 4.19NFGVEE182 pKa = 4.31AFFSFTGVEE191 pKa = 4.01KK192 pKa = 10.1LTLFGGIAFQSEE204 pKa = 4.3DD205 pKa = 4.2DD206 pKa = 4.91DD207 pKa = 4.17PTDD210 pKa = 4.35GYY212 pKa = 9.6TSPEE216 pKa = 3.28IMTFNFWASYY226 pKa = 9.65EE227 pKa = 4.29ISDD230 pKa = 3.35KK231 pKa = 10.81WLVAGEE237 pKa = 4.4FTFSDD242 pKa = 3.29QKK244 pKa = 11.74DD245 pKa = 3.8YY246 pKa = 11.14MDD248 pKa = 4.66GYY250 pKa = 11.4NWLALAKK257 pKa = 10.37YY258 pKa = 10.02AITPKK263 pKa = 9.6ISAVFRR269 pKa = 11.84VSGEE273 pKa = 3.96DD274 pKa = 3.12VDD276 pKa = 4.8YY277 pKa = 10.92EE278 pKa = 4.59DD279 pKa = 6.15PIEE282 pKa = 4.29TDD284 pKa = 3.14PSFLKK289 pKa = 9.88FTLAPAYY296 pKa = 8.68TITDD300 pKa = 3.79NLSVTAEE307 pKa = 3.84VSYY310 pKa = 11.43YY311 pKa = 10.54DD312 pKa = 3.59YY313 pKa = 11.33KK314 pKa = 11.48DD315 pKa = 3.69YY316 pKa = 11.31VVKK319 pKa = 10.76DD320 pKa = 3.69DD321 pKa = 4.05VFVGVQAIFKK331 pKa = 9.48FF332 pKa = 3.98
Molecular weight: 36.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A178IGB6|A0A178IGB6_9BACT Uncharacterized protein OS=Opitutaceae bacterium TSB47 OX=1184151 GN=AW736_18890 PE=4 SV=1
MM1 pKa = 7.52KK2 pKa = 8.83PTFRR6 pKa = 11.84PHH8 pKa = 5.5RR9 pKa = 11.84KK10 pKa = 9.07KK11 pKa = 10.33RR12 pKa = 11.84VRR14 pKa = 11.84KK15 pKa = 8.27IGFRR19 pKa = 11.84ARR21 pKa = 11.84SATKK25 pKa = 10.17GGRR28 pKa = 11.84KK29 pKa = 9.08VLANRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84VGRR39 pKa = 11.84KK40 pKa = 8.97RR41 pKa = 11.84LTVAA45 pKa = 4.08
MM1 pKa = 7.52KK2 pKa = 8.83PTFRR6 pKa = 11.84PHH8 pKa = 5.5RR9 pKa = 11.84KK10 pKa = 9.07KK11 pKa = 10.33RR12 pKa = 11.84VRR14 pKa = 11.84KK15 pKa = 8.27IGFRR19 pKa = 11.84ARR21 pKa = 11.84SATKK25 pKa = 10.17GGRR28 pKa = 11.84KK29 pKa = 9.08VLANRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84VGRR39 pKa = 11.84KK40 pKa = 8.97RR41 pKa = 11.84LTVAA45 pKa = 4.08
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2146511 |
41 |
16945 |
429.9 |
46.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.216 ± 0.062 | 0.871 ± 0.015 |
5.384 ± 0.023 | 4.915 ± 0.054 |
3.917 ± 0.025 | 9.211 ± 0.117 |
2.105 ± 0.021 | 4.874 ± 0.026 |
3.437 ± 0.044 | 9.632 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.863 ± 0.021 | 3.537 ± 0.045 |
5.235 ± 0.064 | 2.922 ± 0.025 |
6.704 ± 0.065 | 5.878 ± 0.061 |
6.302 ± 0.083 | 6.686 ± 0.028 |
1.624 ± 0.017 | 2.689 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
