
Camel associated porprismacovirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
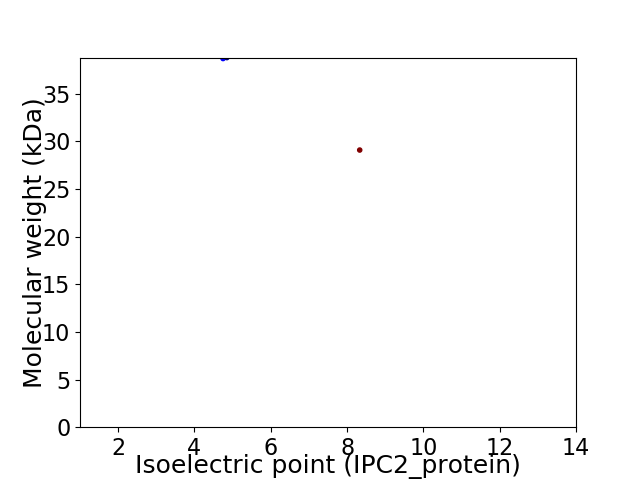
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1EKX0|A0A0A1EKX0_9VIRU Putative replicase protein OS=Camel associated porprismacovirus 4 OX=2170108 GN=rep PE=4 SV=1
MM1 pKa = 7.31ATQTVHH7 pKa = 5.84ATYY10 pKa = 10.67QEE12 pKa = 4.36IYY14 pKa = 10.55DD15 pKa = 4.48LSTKK19 pKa = 10.35PGTLSVIGVHH29 pKa = 5.71TPTGRR34 pKa = 11.84KK35 pKa = 8.95PYY37 pKa = 11.35DD38 pKa = 3.17MLQGFFRR45 pKa = 11.84QFKK48 pKa = 8.4KK49 pKa = 10.75YY50 pKa = 9.66KK51 pKa = 9.06YY52 pKa = 9.92NGITRR57 pKa = 11.84LVLQPAAQLPADD69 pKa = 4.21PLQVSLEE76 pKa = 4.12AGEE79 pKa = 4.62NLDD82 pKa = 5.42PRR84 pKa = 11.84DD85 pKa = 3.89LLNPIMFHH93 pKa = 6.31GAHH96 pKa = 5.75GTDD99 pKa = 3.17INAALNVIYY108 pKa = 10.34KK109 pKa = 10.05KK110 pKa = 10.47QGFNFQTNSTDD121 pKa = 3.78TIDD124 pKa = 4.62LSMFANVEE132 pKa = 4.19GEE134 pKa = 4.29VMDD137 pKa = 4.09SVVVEE142 pKa = 3.86NMYY145 pKa = 11.01YY146 pKa = 10.37SALSDD151 pKa = 3.48PSFRR155 pKa = 11.84KK156 pKa = 10.25YY157 pKa = 10.5GVQEE161 pKa = 4.27VIDD164 pKa = 4.29IGPLVPLVFRR174 pKa = 11.84VNTNMYY180 pKa = 9.66FGPQMSYY187 pKa = 10.75PRR189 pKa = 11.84DD190 pKa = 3.5VFPNVGTSSPNGGFSVSDD208 pKa = 3.42IPSEE212 pKa = 4.13VWNGSSNVSAQTYY225 pKa = 7.22GTDD228 pKa = 3.35MTDD231 pKa = 3.03MQIFTNGVTEE241 pKa = 4.41LSWMPTRR248 pKa = 11.84NAKK251 pKa = 9.75PSSVGTDD258 pKa = 3.53GSPTNTAFVTVPKK271 pKa = 10.12LYY273 pKa = 9.47MGAIIMPPAYY283 pKa = 9.73NQRR286 pKa = 11.84LFYY289 pKa = 10.6RR290 pKa = 11.84CVITHH295 pKa = 6.77SFTFSDD301 pKa = 3.83FGTYY305 pKa = 9.9FDD307 pKa = 4.71GGLNGGSEE315 pKa = 4.57CYY317 pKa = 10.44YY318 pKa = 11.43NNLDD322 pKa = 3.69YY323 pKa = 11.01VAPATMSSGNLDD335 pKa = 3.75SLNLTDD341 pKa = 4.97VEE343 pKa = 4.59GTMRR347 pKa = 11.84TSGVMM352 pKa = 3.2
MM1 pKa = 7.31ATQTVHH7 pKa = 5.84ATYY10 pKa = 10.67QEE12 pKa = 4.36IYY14 pKa = 10.55DD15 pKa = 4.48LSTKK19 pKa = 10.35PGTLSVIGVHH29 pKa = 5.71TPTGRR34 pKa = 11.84KK35 pKa = 8.95PYY37 pKa = 11.35DD38 pKa = 3.17MLQGFFRR45 pKa = 11.84QFKK48 pKa = 8.4KK49 pKa = 10.75YY50 pKa = 9.66KK51 pKa = 9.06YY52 pKa = 9.92NGITRR57 pKa = 11.84LVLQPAAQLPADD69 pKa = 4.21PLQVSLEE76 pKa = 4.12AGEE79 pKa = 4.62NLDD82 pKa = 5.42PRR84 pKa = 11.84DD85 pKa = 3.89LLNPIMFHH93 pKa = 6.31GAHH96 pKa = 5.75GTDD99 pKa = 3.17INAALNVIYY108 pKa = 10.34KK109 pKa = 10.05KK110 pKa = 10.47QGFNFQTNSTDD121 pKa = 3.78TIDD124 pKa = 4.62LSMFANVEE132 pKa = 4.19GEE134 pKa = 4.29VMDD137 pKa = 4.09SVVVEE142 pKa = 3.86NMYY145 pKa = 11.01YY146 pKa = 10.37SALSDD151 pKa = 3.48PSFRR155 pKa = 11.84KK156 pKa = 10.25YY157 pKa = 10.5GVQEE161 pKa = 4.27VIDD164 pKa = 4.29IGPLVPLVFRR174 pKa = 11.84VNTNMYY180 pKa = 9.66FGPQMSYY187 pKa = 10.75PRR189 pKa = 11.84DD190 pKa = 3.5VFPNVGTSSPNGGFSVSDD208 pKa = 3.42IPSEE212 pKa = 4.13VWNGSSNVSAQTYY225 pKa = 7.22GTDD228 pKa = 3.35MTDD231 pKa = 3.03MQIFTNGVTEE241 pKa = 4.41LSWMPTRR248 pKa = 11.84NAKK251 pKa = 9.75PSSVGTDD258 pKa = 3.53GSPTNTAFVTVPKK271 pKa = 10.12LYY273 pKa = 9.47MGAIIMPPAYY283 pKa = 9.73NQRR286 pKa = 11.84LFYY289 pKa = 10.6RR290 pKa = 11.84CVITHH295 pKa = 6.77SFTFSDD301 pKa = 3.83FGTYY305 pKa = 9.9FDD307 pKa = 4.71GGLNGGSEE315 pKa = 4.57CYY317 pKa = 10.44YY318 pKa = 11.43NNLDD322 pKa = 3.69YY323 pKa = 11.01VAPATMSSGNLDD335 pKa = 3.75SLNLTDD341 pKa = 4.97VEE343 pKa = 4.59GTMRR347 pKa = 11.84TSGVMM352 pKa = 3.2
Molecular weight: 38.71 kDa
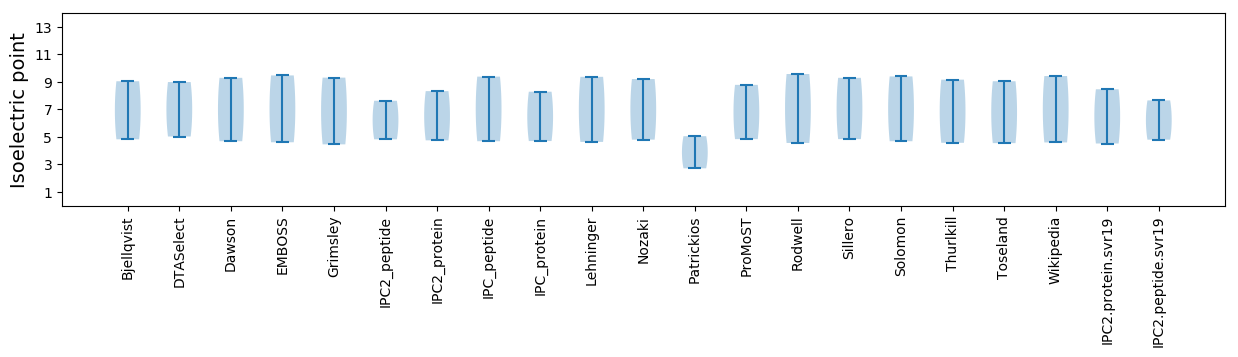
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1EKX0|A0A0A1EKX0_9VIRU Putative replicase protein OS=Camel associated porprismacovirus 4 OX=2170108 GN=rep PE=4 SV=1
MM1 pKa = 7.1LTIPAKK7 pKa = 10.35RR8 pKa = 11.84GEE10 pKa = 3.85FDD12 pKa = 3.79FYY14 pKa = 11.26HH15 pKa = 6.21WLKK18 pKa = 10.88LFEE21 pKa = 3.97VWNVKK26 pKa = 9.5KK27 pKa = 10.12HH28 pKa = 4.32VVGLEE33 pKa = 3.85EE34 pKa = 4.28GKK36 pKa = 10.26NGYY39 pKa = 8.28KK40 pKa = 9.77HH41 pKa = 4.48WQARR45 pKa = 11.84IQLSGEE51 pKa = 4.06GKK53 pKa = 10.12QIIGGSEE60 pKa = 3.41ISFFQYY66 pKa = 10.06MKK68 pKa = 10.0IYY70 pKa = 9.46YY71 pKa = 8.34PKK73 pKa = 10.66AHH75 pKa = 6.86IEE77 pKa = 4.06EE78 pKa = 5.02ASNTWEE84 pKa = 4.12YY85 pKa = 10.62EE86 pKa = 3.99RR87 pKa = 11.84KK88 pKa = 9.62EE89 pKa = 4.23GKK91 pKa = 9.45FWTSEE96 pKa = 3.78DD97 pKa = 3.63TASILAVRR105 pKa = 11.84FGSLRR110 pKa = 11.84PEE112 pKa = 3.91QKK114 pKa = 10.41KK115 pKa = 9.82ILQILEE121 pKa = 4.21SQGDD125 pKa = 3.72RR126 pKa = 11.84EE127 pKa = 4.01IDD129 pKa = 3.26VWLDD133 pKa = 2.99PSGNHH138 pKa = 5.31GKK140 pKa = 9.4SWLTVHH146 pKa = 6.55LWEE149 pKa = 4.48TGRR152 pKa = 11.84ALVVPRR158 pKa = 11.84SSTTAEE164 pKa = 4.04KK165 pKa = 10.66LSAFICSSWKK175 pKa = 10.52GEE177 pKa = 4.2PIVIIDD183 pKa = 3.9IPRR186 pKa = 11.84STKK189 pKa = 10.16VSGSLLEE196 pKa = 4.23TMEE199 pKa = 4.12EE200 pKa = 4.16LKK202 pKa = 11.09DD203 pKa = 3.59GLVFDD208 pKa = 4.17HH209 pKa = 7.47RR210 pKa = 11.84YY211 pKa = 7.63TGRR214 pKa = 11.84TRR216 pKa = 11.84NVRR219 pKa = 11.84GVKK222 pKa = 10.51VMVFTNSEE230 pKa = 4.15LPLKK234 pKa = 10.42KK235 pKa = 10.14LSKK238 pKa = 10.58DD239 pKa = 2.57RR240 pKa = 11.84WRR242 pKa = 11.84LHH244 pKa = 7.0GIACDD249 pKa = 4.01GSLTT253 pKa = 3.83
MM1 pKa = 7.1LTIPAKK7 pKa = 10.35RR8 pKa = 11.84GEE10 pKa = 3.85FDD12 pKa = 3.79FYY14 pKa = 11.26HH15 pKa = 6.21WLKK18 pKa = 10.88LFEE21 pKa = 3.97VWNVKK26 pKa = 9.5KK27 pKa = 10.12HH28 pKa = 4.32VVGLEE33 pKa = 3.85EE34 pKa = 4.28GKK36 pKa = 10.26NGYY39 pKa = 8.28KK40 pKa = 9.77HH41 pKa = 4.48WQARR45 pKa = 11.84IQLSGEE51 pKa = 4.06GKK53 pKa = 10.12QIIGGSEE60 pKa = 3.41ISFFQYY66 pKa = 10.06MKK68 pKa = 10.0IYY70 pKa = 9.46YY71 pKa = 8.34PKK73 pKa = 10.66AHH75 pKa = 6.86IEE77 pKa = 4.06EE78 pKa = 5.02ASNTWEE84 pKa = 4.12YY85 pKa = 10.62EE86 pKa = 3.99RR87 pKa = 11.84KK88 pKa = 9.62EE89 pKa = 4.23GKK91 pKa = 9.45FWTSEE96 pKa = 3.78DD97 pKa = 3.63TASILAVRR105 pKa = 11.84FGSLRR110 pKa = 11.84PEE112 pKa = 3.91QKK114 pKa = 10.41KK115 pKa = 9.82ILQILEE121 pKa = 4.21SQGDD125 pKa = 3.72RR126 pKa = 11.84EE127 pKa = 4.01IDD129 pKa = 3.26VWLDD133 pKa = 2.99PSGNHH138 pKa = 5.31GKK140 pKa = 9.4SWLTVHH146 pKa = 6.55LWEE149 pKa = 4.48TGRR152 pKa = 11.84ALVVPRR158 pKa = 11.84SSTTAEE164 pKa = 4.04KK165 pKa = 10.66LSAFICSSWKK175 pKa = 10.52GEE177 pKa = 4.2PIVIIDD183 pKa = 3.9IPRR186 pKa = 11.84STKK189 pKa = 10.16VSGSLLEE196 pKa = 4.23TMEE199 pKa = 4.12EE200 pKa = 4.16LKK202 pKa = 11.09DD203 pKa = 3.59GLVFDD208 pKa = 4.17HH209 pKa = 7.47RR210 pKa = 11.84YY211 pKa = 7.63TGRR214 pKa = 11.84TRR216 pKa = 11.84NVRR219 pKa = 11.84GVKK222 pKa = 10.51VMVFTNSEE230 pKa = 4.15LPLKK234 pKa = 10.42KK235 pKa = 10.14LSKK238 pKa = 10.58DD239 pKa = 2.57RR240 pKa = 11.84WRR242 pKa = 11.84LHH244 pKa = 7.0GIACDD249 pKa = 4.01GSLTT253 pKa = 3.83
Molecular weight: 29.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
605 |
253 |
352 |
302.5 |
33.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.628 ± 0.414 | 0.661 ± 0.079 |
5.124 ± 0.717 | 5.455 ± 1.985 |
4.793 ± 0.515 | 8.43 ± 0.079 |
2.149 ± 0.62 | 5.124 ± 0.977 |
5.289 ± 2.086 | 7.769 ± 0.81 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.306 ± 1.056 | 4.959 ± 1.584 |
5.124 ± 1.201 | 3.471 ± 0.431 |
4.298 ± 0.999 | 8.264 ± 0.022 |
7.438 ± 1.166 | 7.438 ± 0.682 |
1.983 ± 1.206 | 4.298 ± 0.937 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
