
Kumasi rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ledantevirus; Kumasi ledantevirus
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins

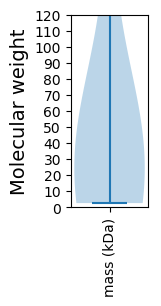
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0C4MKA3|A0A0C4MKA3_9RHAB G OS=Kumasi rhabdovirus OX=1537975 PE=4 SV=1
MM1 pKa = 7.39SFSKK5 pKa = 10.85PEE7 pKa = 3.66AVYY10 pKa = 8.34RR11 pKa = 11.84TCNHH15 pKa = 5.95KK16 pKa = 9.63QVKK19 pKa = 9.01PVLPQEE25 pKa = 4.29TTPIQYY31 pKa = 10.67ASAWFRR37 pKa = 11.84ANRR40 pKa = 11.84ARR42 pKa = 11.84KK43 pKa = 6.4PTLVVGLKK51 pKa = 9.84NVPLVQTLQHH61 pKa = 6.73AAQAMIEE68 pKa = 4.01GRR70 pKa = 11.84LTGDD74 pKa = 3.0IAAATLYY81 pKa = 10.72KK82 pKa = 10.09IFKK85 pKa = 8.95SQKK88 pKa = 9.27QRR90 pKa = 11.84LSLDD94 pKa = 3.1WVSFGVVIGNADD106 pKa = 3.8TEE108 pKa = 4.47VCPWDD113 pKa = 3.9LVNVDD118 pKa = 4.24EE119 pKa = 6.92RR120 pKa = 11.84DD121 pKa = 3.53TQLPNEE127 pKa = 4.53QINDD131 pKa = 3.67QIRR134 pKa = 11.84EE135 pKa = 3.81EE136 pKa = 4.12DD137 pKa = 3.78APWIAFYY144 pKa = 10.61ICFLYY149 pKa = 10.25RR150 pKa = 11.84YY151 pKa = 9.46SKK153 pKa = 9.9ATHH156 pKa = 5.68TDD158 pKa = 3.36YY159 pKa = 11.7RR160 pKa = 11.84DD161 pKa = 3.41MLYY164 pKa = 8.8NTAKK168 pKa = 10.64DD169 pKa = 3.67HH170 pKa = 6.98AKK172 pKa = 10.27HH173 pKa = 5.83LSQRR177 pKa = 11.84AMDD180 pKa = 3.98IQPGSLAQMKK190 pKa = 10.25NVGLDD195 pKa = 3.24ANYY198 pKa = 10.41NCAIACIDD206 pKa = 3.67MFHH209 pKa = 7.04HH210 pKa = 6.74KK211 pKa = 10.15FKK213 pKa = 10.87NLPLSAVRR221 pKa = 11.84YY222 pKa = 6.23GTLPSRR228 pKa = 11.84FKK230 pKa = 10.97DD231 pKa = 3.78CSALTTLNHH240 pKa = 5.35ITKK243 pKa = 8.92LTGLAIEE250 pKa = 5.19QFMLWVFSSKK260 pKa = 10.02MADD263 pKa = 3.83EE264 pKa = 5.24LDD266 pKa = 3.82QMSKK270 pKa = 10.53PDD272 pKa = 3.65EE273 pKa = 4.39EE274 pKa = 5.59LDD276 pKa = 3.62QGDD279 pKa = 4.0SYY281 pKa = 10.48VAYY284 pKa = 9.39MRR286 pKa = 11.84EE287 pKa = 4.09LGVSDD292 pKa = 4.33RR293 pKa = 11.84SPYY296 pKa = 9.8SAQANPSFSLFCHH309 pKa = 5.41VVGALLGSKK318 pKa = 9.96RR319 pKa = 11.84SKK321 pKa = 8.86NARR324 pKa = 11.84MGAEE328 pKa = 3.66VDD330 pKa = 3.91TVNSVLNGKK339 pKa = 8.88IVAYY343 pKa = 10.49VLGTRR348 pKa = 11.84PSFTKK353 pKa = 10.24EE354 pKa = 3.49YY355 pKa = 9.71STDD358 pKa = 3.56KK359 pKa = 11.41EE360 pKa = 4.01EE361 pKa = 5.35DD362 pKa = 3.35PTGTAEE368 pKa = 4.1EE369 pKa = 4.74DD370 pKa = 3.66VTPGEE375 pKa = 4.61IPNGANPDD383 pKa = 2.87EE384 pKa = 4.27WFAYY388 pKa = 10.35LSIKK392 pKa = 10.61GFVLPDD398 pKa = 3.59EE399 pKa = 4.43VVHH402 pKa = 5.27WVKK405 pKa = 9.91TRR407 pKa = 11.84VQLLTDD413 pKa = 3.46VRR415 pKa = 11.84DD416 pKa = 3.81GTVGKK421 pKa = 9.62FLKK424 pKa = 9.27TQGG427 pKa = 3.4
MM1 pKa = 7.39SFSKK5 pKa = 10.85PEE7 pKa = 3.66AVYY10 pKa = 8.34RR11 pKa = 11.84TCNHH15 pKa = 5.95KK16 pKa = 9.63QVKK19 pKa = 9.01PVLPQEE25 pKa = 4.29TTPIQYY31 pKa = 10.67ASAWFRR37 pKa = 11.84ANRR40 pKa = 11.84ARR42 pKa = 11.84KK43 pKa = 6.4PTLVVGLKK51 pKa = 9.84NVPLVQTLQHH61 pKa = 6.73AAQAMIEE68 pKa = 4.01GRR70 pKa = 11.84LTGDD74 pKa = 3.0IAAATLYY81 pKa = 10.72KK82 pKa = 10.09IFKK85 pKa = 8.95SQKK88 pKa = 9.27QRR90 pKa = 11.84LSLDD94 pKa = 3.1WVSFGVVIGNADD106 pKa = 3.8TEE108 pKa = 4.47VCPWDD113 pKa = 3.9LVNVDD118 pKa = 4.24EE119 pKa = 6.92RR120 pKa = 11.84DD121 pKa = 3.53TQLPNEE127 pKa = 4.53QINDD131 pKa = 3.67QIRR134 pKa = 11.84EE135 pKa = 3.81EE136 pKa = 4.12DD137 pKa = 3.78APWIAFYY144 pKa = 10.61ICFLYY149 pKa = 10.25RR150 pKa = 11.84YY151 pKa = 9.46SKK153 pKa = 9.9ATHH156 pKa = 5.68TDD158 pKa = 3.36YY159 pKa = 11.7RR160 pKa = 11.84DD161 pKa = 3.41MLYY164 pKa = 8.8NTAKK168 pKa = 10.64DD169 pKa = 3.67HH170 pKa = 6.98AKK172 pKa = 10.27HH173 pKa = 5.83LSQRR177 pKa = 11.84AMDD180 pKa = 3.98IQPGSLAQMKK190 pKa = 10.25NVGLDD195 pKa = 3.24ANYY198 pKa = 10.41NCAIACIDD206 pKa = 3.67MFHH209 pKa = 7.04HH210 pKa = 6.74KK211 pKa = 10.15FKK213 pKa = 10.87NLPLSAVRR221 pKa = 11.84YY222 pKa = 6.23GTLPSRR228 pKa = 11.84FKK230 pKa = 10.97DD231 pKa = 3.78CSALTTLNHH240 pKa = 5.35ITKK243 pKa = 8.92LTGLAIEE250 pKa = 5.19QFMLWVFSSKK260 pKa = 10.02MADD263 pKa = 3.83EE264 pKa = 5.24LDD266 pKa = 3.82QMSKK270 pKa = 10.53PDD272 pKa = 3.65EE273 pKa = 4.39EE274 pKa = 5.59LDD276 pKa = 3.62QGDD279 pKa = 4.0SYY281 pKa = 10.48VAYY284 pKa = 9.39MRR286 pKa = 11.84EE287 pKa = 4.09LGVSDD292 pKa = 4.33RR293 pKa = 11.84SPYY296 pKa = 9.8SAQANPSFSLFCHH309 pKa = 5.41VVGALLGSKK318 pKa = 9.96RR319 pKa = 11.84SKK321 pKa = 8.86NARR324 pKa = 11.84MGAEE328 pKa = 3.66VDD330 pKa = 3.91TVNSVLNGKK339 pKa = 8.88IVAYY343 pKa = 10.49VLGTRR348 pKa = 11.84PSFTKK353 pKa = 10.24EE354 pKa = 3.49YY355 pKa = 9.71STDD358 pKa = 3.56KK359 pKa = 11.41EE360 pKa = 4.01EE361 pKa = 5.35DD362 pKa = 3.35PTGTAEE368 pKa = 4.1EE369 pKa = 4.74DD370 pKa = 3.66VTPGEE375 pKa = 4.61IPNGANPDD383 pKa = 2.87EE384 pKa = 4.27WFAYY388 pKa = 10.35LSIKK392 pKa = 10.61GFVLPDD398 pKa = 3.59EE399 pKa = 4.43VVHH402 pKa = 5.27WVKK405 pKa = 9.91TRR407 pKa = 11.84VQLLTDD413 pKa = 3.46VRR415 pKa = 11.84DD416 pKa = 3.81GTVGKK421 pKa = 9.62FLKK424 pKa = 9.27TQGG427 pKa = 3.4
Molecular weight: 47.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C4ME19|A0A0C4ME19_9RHAB GDP polyribonucleotidyltransferase OS=Kumasi rhabdovirus OX=1537975 PE=4 SV=1
MM1 pKa = 8.03DD2 pKa = 4.39KK3 pKa = 10.35MKK5 pKa = 10.92SSLLAKK11 pKa = 9.54MMKK14 pKa = 10.53KK15 pKa = 9.73MMTEE19 pKa = 3.81EE20 pKa = 3.95VEE22 pKa = 3.97MRR24 pKa = 11.84ILGLIPYY31 pKa = 9.39LKK33 pKa = 9.72TPNSLEE39 pKa = 4.43GDD41 pKa = 3.27WKK43 pKa = 7.87MTRR46 pKa = 11.84PEE48 pKa = 3.86RR49 pKa = 11.84RR50 pKa = 11.84LGRR53 pKa = 11.84YY54 pKa = 7.67HH55 pKa = 5.42WRR57 pKa = 11.84KK58 pKa = 9.23RR59 pKa = 11.84VNQLRR64 pKa = 11.84YY65 pKa = 9.5RR66 pKa = 11.84VFLLRR71 pKa = 11.84LL72 pKa = 3.53
MM1 pKa = 8.03DD2 pKa = 4.39KK3 pKa = 10.35MKK5 pKa = 10.92SSLLAKK11 pKa = 9.54MMKK14 pKa = 10.53KK15 pKa = 9.73MMTEE19 pKa = 3.81EE20 pKa = 3.95VEE22 pKa = 3.97MRR24 pKa = 11.84ILGLIPYY31 pKa = 9.39LKK33 pKa = 9.72TPNSLEE39 pKa = 4.43GDD41 pKa = 3.27WKK43 pKa = 7.87MTRR46 pKa = 11.84PEE48 pKa = 3.86RR49 pKa = 11.84RR50 pKa = 11.84LGRR53 pKa = 11.84YY54 pKa = 7.67HH55 pKa = 5.42WRR57 pKa = 11.84KK58 pKa = 9.23RR59 pKa = 11.84VNQLRR64 pKa = 11.84YY65 pKa = 9.5RR66 pKa = 11.84VFLLRR71 pKa = 11.84LL72 pKa = 3.53
Molecular weight: 8.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3458 |
22 |
2122 |
494.0 |
56.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.685 ± 0.921 | 2.371 ± 0.219 |
5.234 ± 0.348 | 5.726 ± 0.173 |
4.078 ± 0.321 | 6.333 ± 0.515 |
3.036 ± 0.378 | 6.073 ± 0.653 |
6.333 ± 0.484 | 9.746 ± 1.226 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.429 ± 0.33 | 4.627 ± 0.49 |
4.858 ± 0.477 | 3.152 ± 0.365 |
5.552 ± 0.642 | 7.577 ± 0.869 |
6.189 ± 0.43 | 6.391 ± 0.901 |
2.198 ± 0.286 | 3.412 ± 0.201 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
