
Oat golden stripe virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Furovirus
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
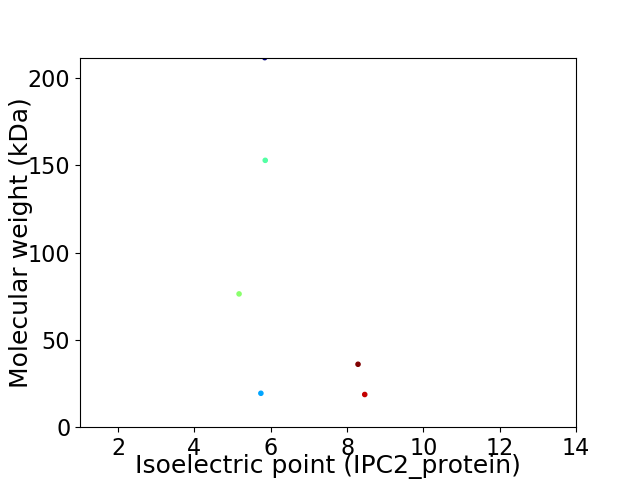
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
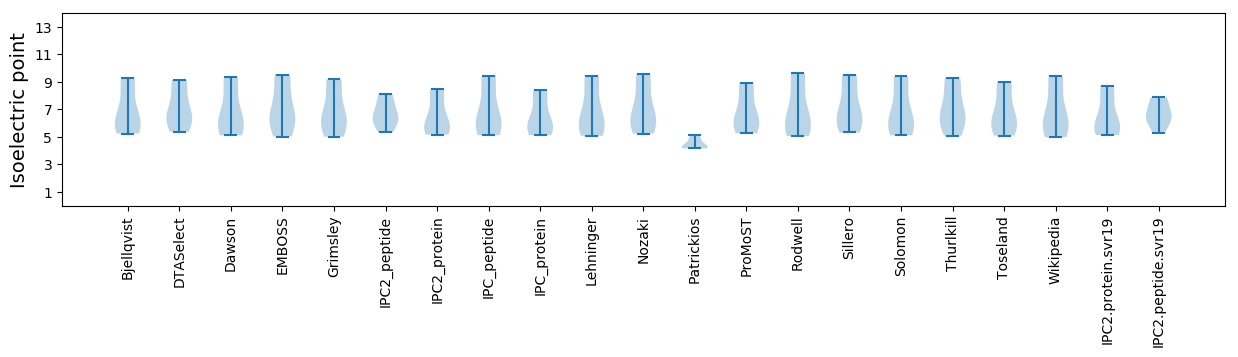
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9IWA9|Q9IWA9_9VIRU Methyltransferase/RNA helicase OS=Oat golden stripe virus OX=45103 GN=ORF1 readthrough PE=4 SV=1
MM1 pKa = 7.74PVTPGYY7 pKa = 8.96TGYY10 pKa = 10.93NKK12 pKa = 10.06EE13 pKa = 4.36LNVMANTHH21 pKa = 5.47AFIRR25 pKa = 11.84MSTLMNQIEE34 pKa = 4.87GWQSTRR40 pKa = 11.84ASVLSHH46 pKa = 6.57LGIMLNAVSKK56 pKa = 10.73LGEE59 pKa = 3.92RR60 pKa = 11.84NFFSRR65 pKa = 11.84NKK67 pKa = 10.33RR68 pKa = 11.84FGTHH72 pKa = 5.06TQDD75 pKa = 3.63GDD77 pKa = 4.7EE78 pKa = 4.48IFCDD82 pKa = 3.64LGGEE86 pKa = 4.24AVGQVLGRR94 pKa = 11.84LTVALQPAKK103 pKa = 11.03GEE105 pKa = 4.22GTQTRR110 pKa = 11.84NSKK113 pKa = 10.51RR114 pKa = 11.84GVPPAAGQVEE124 pKa = 4.56GEE126 pKa = 4.27EE127 pKa = 4.43QGQSDD132 pKa = 4.05QTLQIANAVSDD143 pKa = 3.25LMNYY147 pKa = 8.81VRR149 pKa = 11.84TKK151 pKa = 10.59DD152 pKa = 3.42YY153 pKa = 11.4TMNEE157 pKa = 5.24CYY159 pKa = 9.66TQDD162 pKa = 3.5SFEE165 pKa = 4.41AKK167 pKa = 10.55YY168 pKa = 9.68NLKK171 pKa = 10.11WEE173 pKa = 4.69GSAXRR178 pKa = 11.84GGVSEE183 pKa = 4.12KK184 pKa = 10.29TRR186 pKa = 11.84VQVVEE191 pKa = 4.31EE192 pKa = 3.92LADD195 pKa = 4.99RR196 pKa = 11.84IVLADD201 pKa = 4.23DD202 pKa = 5.0LGIFPRR208 pKa = 11.84RR209 pKa = 11.84GEE211 pKa = 3.86NGFIIVDD218 pKa = 3.92EE219 pKa = 4.65PEE221 pKa = 3.78VLYY224 pKa = 11.17AEE226 pKa = 5.11LEE228 pKa = 4.18DD229 pKa = 4.95KK230 pKa = 10.72ILSFEE235 pKa = 4.11HH236 pKa = 6.28SVYY239 pKa = 10.31HH240 pKa = 5.93PRR242 pKa = 11.84AITVEE247 pKa = 4.34TEE249 pKa = 4.01VQPLTVKK256 pKa = 9.62TADD259 pKa = 3.67SEE261 pKa = 4.56NLVMMDD267 pKa = 4.27EE268 pKa = 4.41EE269 pKa = 4.79DD270 pKa = 4.07SIIVTEE276 pKa = 4.04VLVDD280 pKa = 4.06DD281 pKa = 4.58EE282 pKa = 6.04SDD284 pKa = 4.1YY285 pKa = 11.4FWLYY289 pKa = 11.45LLLLLVSVLISLFILLSRR307 pKa = 11.84PFILSLGNRR316 pKa = 11.84RR317 pKa = 11.84EE318 pKa = 3.99VFGGDD323 pKa = 2.62ICYY326 pKa = 10.39KK327 pKa = 8.7IAEE330 pKa = 4.51WLWKK334 pKa = 9.82VRR336 pKa = 11.84MWLLKK341 pKa = 9.48RR342 pKa = 11.84AMYY345 pKa = 10.43RR346 pKa = 11.84FGNLGVVRR354 pKa = 11.84KK355 pKa = 9.1NHH357 pKa = 5.91TKK359 pKa = 10.18SAMRR363 pKa = 11.84RR364 pKa = 11.84YY365 pKa = 9.99LRR367 pKa = 11.84DD368 pKa = 3.15YY369 pKa = 11.06NFQLEE374 pKa = 4.31GWEE377 pKa = 4.1TAINCSPGEE386 pKa = 3.98ALIAADD392 pKa = 3.29QHH394 pKa = 6.4VLVQEE399 pKa = 4.15VRR401 pKa = 11.84EE402 pKa = 4.02LLDD405 pKa = 3.79LEE407 pKa = 4.7GNLMSVEE414 pKa = 4.42KK415 pKa = 10.14IRR417 pKa = 11.84KK418 pKa = 8.8IFEE421 pKa = 4.12NYY423 pKa = 8.97NVNNYY428 pKa = 10.2KK429 pKa = 8.08ITKK432 pKa = 8.7WLDD435 pKa = 3.14KK436 pKa = 10.71QVIRR440 pKa = 11.84HH441 pKa = 6.06ALLAVNEE448 pKa = 4.26TLKK451 pKa = 10.89NDD453 pKa = 4.0EE454 pKa = 4.11ITLEE458 pKa = 3.95EE459 pKa = 4.38GMLNLACATFDD470 pKa = 4.59QILEE474 pKa = 4.38CDD476 pKa = 3.56VEE478 pKa = 4.48TQRR481 pKa = 11.84GSMYY485 pKa = 10.59FGDD488 pKa = 5.58LIALQFAFRR497 pKa = 11.84LIGTPEE503 pKa = 3.65FSDD506 pKa = 3.98FVVAYY511 pKa = 9.55KK512 pKa = 11.1GNLYY516 pKa = 9.45PCYY519 pKa = 9.75VEE521 pKa = 4.8AIKK524 pKa = 10.7RR525 pKa = 11.84HH526 pKa = 4.7ASKK529 pKa = 11.24LEE531 pKa = 3.9LQHH534 pKa = 8.41AEE536 pKa = 3.69DD537 pKa = 4.95DD538 pKa = 3.73DD539 pKa = 4.54RR540 pKa = 11.84KK541 pKa = 10.33RR542 pKa = 11.84SEE544 pKa = 5.09KK545 pKa = 10.58ALEE548 pKa = 3.96WLLMAGQGAFALSSAMEE565 pKa = 4.32TFTPAEE571 pKa = 4.2FQDD574 pKa = 3.45EE575 pKa = 4.25MRR577 pKa = 11.84QVVQAYY583 pKa = 10.4ADD585 pKa = 4.0TTFHH589 pKa = 6.38EE590 pKa = 4.72ADD592 pKa = 3.92RR593 pKa = 11.84PLLSGLGASSIGDD606 pKa = 3.44VASEE610 pKa = 4.24FVTGDD615 pKa = 3.01ISAGTDD621 pKa = 3.26VEE623 pKa = 4.76TSSSLSIGTASRR635 pKa = 11.84ARR637 pKa = 11.84KK638 pKa = 9.47RR639 pKa = 11.84SIARR643 pKa = 11.84NIRR646 pKa = 11.84MYY648 pKa = 10.65PGGAVAHH655 pKa = 6.55GGVKK659 pKa = 10.23APRR662 pKa = 11.84VSGTSGVGGMKK673 pKa = 9.58MKK675 pKa = 9.59FTSKK679 pKa = 10.3PMASS683 pKa = 3.38
MM1 pKa = 7.74PVTPGYY7 pKa = 8.96TGYY10 pKa = 10.93NKK12 pKa = 10.06EE13 pKa = 4.36LNVMANTHH21 pKa = 5.47AFIRR25 pKa = 11.84MSTLMNQIEE34 pKa = 4.87GWQSTRR40 pKa = 11.84ASVLSHH46 pKa = 6.57LGIMLNAVSKK56 pKa = 10.73LGEE59 pKa = 3.92RR60 pKa = 11.84NFFSRR65 pKa = 11.84NKK67 pKa = 10.33RR68 pKa = 11.84FGTHH72 pKa = 5.06TQDD75 pKa = 3.63GDD77 pKa = 4.7EE78 pKa = 4.48IFCDD82 pKa = 3.64LGGEE86 pKa = 4.24AVGQVLGRR94 pKa = 11.84LTVALQPAKK103 pKa = 11.03GEE105 pKa = 4.22GTQTRR110 pKa = 11.84NSKK113 pKa = 10.51RR114 pKa = 11.84GVPPAAGQVEE124 pKa = 4.56GEE126 pKa = 4.27EE127 pKa = 4.43QGQSDD132 pKa = 4.05QTLQIANAVSDD143 pKa = 3.25LMNYY147 pKa = 8.81VRR149 pKa = 11.84TKK151 pKa = 10.59DD152 pKa = 3.42YY153 pKa = 11.4TMNEE157 pKa = 5.24CYY159 pKa = 9.66TQDD162 pKa = 3.5SFEE165 pKa = 4.41AKK167 pKa = 10.55YY168 pKa = 9.68NLKK171 pKa = 10.11WEE173 pKa = 4.69GSAXRR178 pKa = 11.84GGVSEE183 pKa = 4.12KK184 pKa = 10.29TRR186 pKa = 11.84VQVVEE191 pKa = 4.31EE192 pKa = 3.92LADD195 pKa = 4.99RR196 pKa = 11.84IVLADD201 pKa = 4.23DD202 pKa = 5.0LGIFPRR208 pKa = 11.84RR209 pKa = 11.84GEE211 pKa = 3.86NGFIIVDD218 pKa = 3.92EE219 pKa = 4.65PEE221 pKa = 3.78VLYY224 pKa = 11.17AEE226 pKa = 5.11LEE228 pKa = 4.18DD229 pKa = 4.95KK230 pKa = 10.72ILSFEE235 pKa = 4.11HH236 pKa = 6.28SVYY239 pKa = 10.31HH240 pKa = 5.93PRR242 pKa = 11.84AITVEE247 pKa = 4.34TEE249 pKa = 4.01VQPLTVKK256 pKa = 9.62TADD259 pKa = 3.67SEE261 pKa = 4.56NLVMMDD267 pKa = 4.27EE268 pKa = 4.41EE269 pKa = 4.79DD270 pKa = 4.07SIIVTEE276 pKa = 4.04VLVDD280 pKa = 4.06DD281 pKa = 4.58EE282 pKa = 6.04SDD284 pKa = 4.1YY285 pKa = 11.4FWLYY289 pKa = 11.45LLLLLVSVLISLFILLSRR307 pKa = 11.84PFILSLGNRR316 pKa = 11.84RR317 pKa = 11.84EE318 pKa = 3.99VFGGDD323 pKa = 2.62ICYY326 pKa = 10.39KK327 pKa = 8.7IAEE330 pKa = 4.51WLWKK334 pKa = 9.82VRR336 pKa = 11.84MWLLKK341 pKa = 9.48RR342 pKa = 11.84AMYY345 pKa = 10.43RR346 pKa = 11.84FGNLGVVRR354 pKa = 11.84KK355 pKa = 9.1NHH357 pKa = 5.91TKK359 pKa = 10.18SAMRR363 pKa = 11.84RR364 pKa = 11.84YY365 pKa = 9.99LRR367 pKa = 11.84DD368 pKa = 3.15YY369 pKa = 11.06NFQLEE374 pKa = 4.31GWEE377 pKa = 4.1TAINCSPGEE386 pKa = 3.98ALIAADD392 pKa = 3.29QHH394 pKa = 6.4VLVQEE399 pKa = 4.15VRR401 pKa = 11.84EE402 pKa = 4.02LLDD405 pKa = 3.79LEE407 pKa = 4.7GNLMSVEE414 pKa = 4.42KK415 pKa = 10.14IRR417 pKa = 11.84KK418 pKa = 8.8IFEE421 pKa = 4.12NYY423 pKa = 8.97NVNNYY428 pKa = 10.2KK429 pKa = 8.08ITKK432 pKa = 8.7WLDD435 pKa = 3.14KK436 pKa = 10.71QVIRR440 pKa = 11.84HH441 pKa = 6.06ALLAVNEE448 pKa = 4.26TLKK451 pKa = 10.89NDD453 pKa = 4.0EE454 pKa = 4.11ITLEE458 pKa = 3.95EE459 pKa = 4.38GMLNLACATFDD470 pKa = 4.59QILEE474 pKa = 4.38CDD476 pKa = 3.56VEE478 pKa = 4.48TQRR481 pKa = 11.84GSMYY485 pKa = 10.59FGDD488 pKa = 5.58LIALQFAFRR497 pKa = 11.84LIGTPEE503 pKa = 3.65FSDD506 pKa = 3.98FVVAYY511 pKa = 9.55KK512 pKa = 11.1GNLYY516 pKa = 9.45PCYY519 pKa = 9.75VEE521 pKa = 4.8AIKK524 pKa = 10.7RR525 pKa = 11.84HH526 pKa = 4.7ASKK529 pKa = 11.24LEE531 pKa = 3.9LQHH534 pKa = 8.41AEE536 pKa = 3.69DD537 pKa = 4.95DD538 pKa = 3.73DD539 pKa = 4.54RR540 pKa = 11.84KK541 pKa = 10.33RR542 pKa = 11.84SEE544 pKa = 5.09KK545 pKa = 10.58ALEE548 pKa = 3.96WLLMAGQGAFALSSAMEE565 pKa = 4.32TFTPAEE571 pKa = 4.2FQDD574 pKa = 3.45EE575 pKa = 4.25MRR577 pKa = 11.84QVVQAYY583 pKa = 10.4ADD585 pKa = 4.0TTFHH589 pKa = 6.38EE590 pKa = 4.72ADD592 pKa = 3.92RR593 pKa = 11.84PLLSGLGASSIGDD606 pKa = 3.44VASEE610 pKa = 4.24FVTGDD615 pKa = 3.01ISAGTDD621 pKa = 3.26VEE623 pKa = 4.76TSSSLSIGTASRR635 pKa = 11.84ARR637 pKa = 11.84KK638 pKa = 9.47RR639 pKa = 11.84SIARR643 pKa = 11.84NIRR646 pKa = 11.84MYY648 pKa = 10.65PGGAVAHH655 pKa = 6.55GGVKK659 pKa = 10.23APRR662 pKa = 11.84VSGTSGVGGMKK673 pKa = 9.58MKK675 pKa = 9.59FTSKK679 pKa = 10.3PMASS683 pKa = 3.38
Molecular weight: 76.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9QBU6|Q9QBU6_9VIRU Coat protein OS=Oat golden stripe virus OX=45103 GN=cp PE=4 SV=1
MM1 pKa = 7.16SAAVHH6 pKa = 6.07SCAKK10 pKa = 10.06CADD13 pKa = 3.2GFTRR17 pKa = 11.84VVCVSKK23 pKa = 10.98YY24 pKa = 9.91RR25 pKa = 11.84NSVYY29 pKa = 10.41KK30 pKa = 10.2SLGMEE35 pKa = 4.1VVKK38 pKa = 10.78CRR40 pKa = 11.84LPSACGVNCRR50 pKa = 11.84MPAALVLVKK59 pKa = 9.35GHH61 pKa = 7.49PEE63 pKa = 3.45LSMDD67 pKa = 4.04GFCGEE72 pKa = 3.89KK73 pKa = 9.85HH74 pKa = 5.94RR75 pKa = 11.84GYY77 pKa = 10.45VVSGAWRR84 pKa = 11.84HH85 pKa = 4.7AQLRR89 pKa = 11.84SLNAEE94 pKa = 3.95LDD96 pKa = 3.46KK97 pKa = 10.99LTEE100 pKa = 4.26KK101 pKa = 10.91EE102 pKa = 3.89EE103 pKa = 4.15SLRR106 pKa = 11.84TQIIVLTAAAKK117 pKa = 8.81TADD120 pKa = 3.47APVYY124 pKa = 10.38VPKK127 pKa = 10.62KK128 pKa = 9.39LNKK131 pKa = 10.25LKK133 pKa = 11.17VEE135 pKa = 4.15VSDD138 pKa = 3.65VTEE141 pKa = 4.95KK142 pKa = 10.28IHH144 pKa = 6.08QKK146 pKa = 10.34SVGLAVVMDD155 pKa = 5.86AILADD160 pKa = 4.56LSPDD164 pKa = 3.23ASPKK168 pKa = 9.82RR169 pKa = 11.84RR170 pKa = 11.84SPAA173 pKa = 3.07
MM1 pKa = 7.16SAAVHH6 pKa = 6.07SCAKK10 pKa = 10.06CADD13 pKa = 3.2GFTRR17 pKa = 11.84VVCVSKK23 pKa = 10.98YY24 pKa = 9.91RR25 pKa = 11.84NSVYY29 pKa = 10.41KK30 pKa = 10.2SLGMEE35 pKa = 4.1VVKK38 pKa = 10.78CRR40 pKa = 11.84LPSACGVNCRR50 pKa = 11.84MPAALVLVKK59 pKa = 9.35GHH61 pKa = 7.49PEE63 pKa = 3.45LSMDD67 pKa = 4.04GFCGEE72 pKa = 3.89KK73 pKa = 9.85HH74 pKa = 5.94RR75 pKa = 11.84GYY77 pKa = 10.45VVSGAWRR84 pKa = 11.84HH85 pKa = 4.7AQLRR89 pKa = 11.84SLNAEE94 pKa = 3.95LDD96 pKa = 3.46KK97 pKa = 10.99LTEE100 pKa = 4.26KK101 pKa = 10.91EE102 pKa = 3.89EE103 pKa = 4.15SLRR106 pKa = 11.84TQIIVLTAAAKK117 pKa = 8.81TADD120 pKa = 3.47APVYY124 pKa = 10.38VPKK127 pKa = 10.62KK128 pKa = 9.39LNKK131 pKa = 10.25LKK133 pKa = 11.17VEE135 pKa = 4.15VSDD138 pKa = 3.65VTEE141 pKa = 4.95KK142 pKa = 10.28IHH144 pKa = 6.08QKK146 pKa = 10.34SVGLAVVMDD155 pKa = 5.86AILADD160 pKa = 4.56LSPDD164 pKa = 3.23ASPKK168 pKa = 9.82RR169 pKa = 11.84RR170 pKa = 11.84SPAA173 pKa = 3.07
Molecular weight: 18.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4554 |
173 |
1853 |
759.0 |
85.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.851 ± 0.722 | 1.779 ± 0.235 |
6.588 ± 0.478 | 7.312 ± 0.436 |
3.931 ± 0.341 | 5.336 ± 0.694 |
1.954 ± 0.17 | 4.458 ± 0.231 |
7.246 ± 0.65 | 9.025 ± 0.199 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.942 ± 0.227 | 4.414 ± 0.371 |
3.491 ± 0.274 | 3.118 ± 0.305 |
5.797 ± 0.154 | 6.697 ± 0.355 |
5.951 ± 0.308 | 8.608 ± 0.376 |
1.098 ± 0.109 | 3.36 ± 0.095 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
