
Holcus lanatus-associated virus
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses
Average proteome isoelectric point is 8.93
Get precalculated fractions of proteins
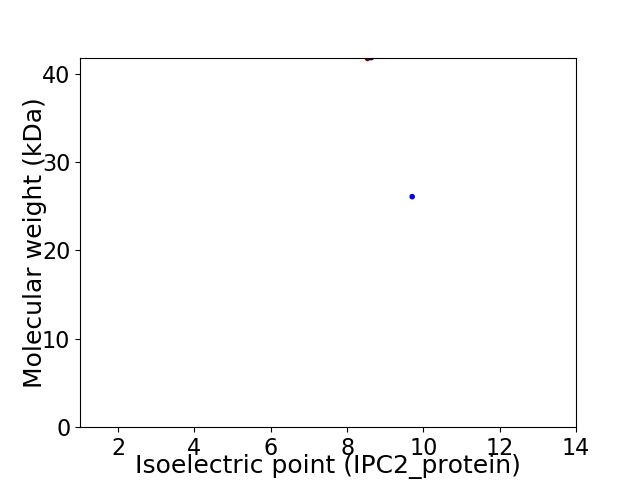
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345BKB0|A0A345BKB0_9VIRU Coat protein OS=Holcus lanatus-associated virus OX=2282643 PE=4 SV=1
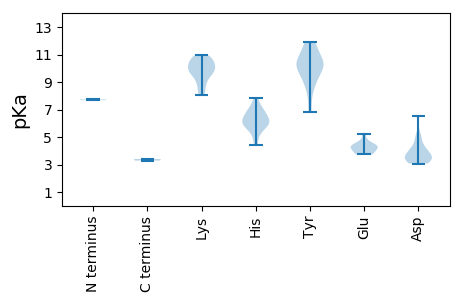
MM1 pKa = 7.73PRR3 pKa = 11.84QKK5 pKa = 10.77SPAHH9 pKa = 6.29DD10 pKa = 3.07WCFTINNPRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84DD22 pKa = 3.52HH23 pKa = 7.13QRR25 pKa = 11.84VQDD28 pKa = 3.3WQYY31 pKa = 11.76NYY33 pKa = 10.7LIYY36 pKa = 10.37QLEE39 pKa = 4.43VGDD42 pKa = 3.83QGTPHH47 pKa = 6.52LQGFVQFTTKK57 pKa = 9.92QRR59 pKa = 11.84LTALKK64 pKa = 9.67KK65 pKa = 9.51LHH67 pKa = 6.89KK68 pKa = 9.34SAHH71 pKa = 5.16WEE73 pKa = 3.76PRR75 pKa = 11.84RR76 pKa = 11.84GSAYY80 pKa = 9.8QASHH84 pKa = 5.95YY85 pKa = 10.01CEE87 pKa = 5.25KK88 pKa = 10.49PVPDD92 pKa = 5.09CDD94 pKa = 3.59CHH96 pKa = 7.87HH97 pKa = 6.89CVDD100 pKa = 4.07TPRR103 pKa = 11.84VYY105 pKa = 10.67PQFIFKK111 pKa = 10.18NGNISAPAGEE121 pKa = 4.77KK122 pKa = 8.95LTSIARR128 pKa = 11.84VIKK131 pKa = 9.99TKK133 pKa = 10.56GLSAAIDD140 pKa = 3.95NYY142 pKa = 7.55PTHH145 pKa = 6.72YY146 pKa = 10.01MGMNRR151 pKa = 11.84GMVNRR156 pKa = 11.84PPINYY161 pKa = 9.3KK162 pKa = 9.83EE163 pKa = 3.99ALANFYY169 pKa = 10.86SPQRR173 pKa = 11.84DD174 pKa = 3.24WQTVVSICYY183 pKa = 8.81GKK185 pKa = 9.9PEE187 pKa = 4.19TGKK190 pKa = 8.24TRR192 pKa = 11.84YY193 pKa = 10.36AMLAPSPYY201 pKa = 10.26KK202 pKa = 10.41LAAFGAEE209 pKa = 3.74GSTDD213 pKa = 3.55FFGDD217 pKa = 3.56YY218 pKa = 10.57RR219 pKa = 11.84PDD221 pKa = 3.18QHH223 pKa = 6.04EE224 pKa = 4.36TLVVDD229 pKa = 5.11DD230 pKa = 6.51FYY232 pKa = 11.94SNWKK236 pKa = 8.04YY237 pKa = 6.84TTFLQVCDD245 pKa = 4.41RR246 pKa = 11.84YY247 pKa = 8.69PTEE250 pKa = 3.81VHH252 pKa = 5.73TKK254 pKa = 9.97GGFRR258 pKa = 11.84QLLVRR263 pKa = 11.84HH264 pKa = 6.0IVFTSNHH271 pKa = 5.91SPGVWYY277 pKa = 9.29PNVLADD283 pKa = 3.61ADD285 pKa = 3.91RR286 pKa = 11.84RR287 pKa = 11.84EE288 pKa = 4.18SFNRR292 pKa = 11.84RR293 pKa = 11.84IHH295 pKa = 5.9NIIWFTPDD303 pKa = 3.27GYY305 pKa = 9.39WVKK308 pKa = 10.65KK309 pKa = 8.81GTLPWPCPFLQPLAWNHH326 pKa = 4.4VLRR329 pKa = 11.84NEE331 pKa = 4.11MLPIALLPQNQNLTIHH347 pKa = 6.18EE348 pKa = 4.46RR349 pKa = 11.84GILDD353 pKa = 4.06PSQLFVV359 pKa = 3.4
MM1 pKa = 7.73PRR3 pKa = 11.84QKK5 pKa = 10.77SPAHH9 pKa = 6.29DD10 pKa = 3.07WCFTINNPRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84DD22 pKa = 3.52HH23 pKa = 7.13QRR25 pKa = 11.84VQDD28 pKa = 3.3WQYY31 pKa = 11.76NYY33 pKa = 10.7LIYY36 pKa = 10.37QLEE39 pKa = 4.43VGDD42 pKa = 3.83QGTPHH47 pKa = 6.52LQGFVQFTTKK57 pKa = 9.92QRR59 pKa = 11.84LTALKK64 pKa = 9.67KK65 pKa = 9.51LHH67 pKa = 6.89KK68 pKa = 9.34SAHH71 pKa = 5.16WEE73 pKa = 3.76PRR75 pKa = 11.84RR76 pKa = 11.84GSAYY80 pKa = 9.8QASHH84 pKa = 5.95YY85 pKa = 10.01CEE87 pKa = 5.25KK88 pKa = 10.49PVPDD92 pKa = 5.09CDD94 pKa = 3.59CHH96 pKa = 7.87HH97 pKa = 6.89CVDD100 pKa = 4.07TPRR103 pKa = 11.84VYY105 pKa = 10.67PQFIFKK111 pKa = 10.18NGNISAPAGEE121 pKa = 4.77KK122 pKa = 8.95LTSIARR128 pKa = 11.84VIKK131 pKa = 9.99TKK133 pKa = 10.56GLSAAIDD140 pKa = 3.95NYY142 pKa = 7.55PTHH145 pKa = 6.72YY146 pKa = 10.01MGMNRR151 pKa = 11.84GMVNRR156 pKa = 11.84PPINYY161 pKa = 9.3KK162 pKa = 9.83EE163 pKa = 3.99ALANFYY169 pKa = 10.86SPQRR173 pKa = 11.84DD174 pKa = 3.24WQTVVSICYY183 pKa = 8.81GKK185 pKa = 9.9PEE187 pKa = 4.19TGKK190 pKa = 8.24TRR192 pKa = 11.84YY193 pKa = 10.36AMLAPSPYY201 pKa = 10.26KK202 pKa = 10.41LAAFGAEE209 pKa = 3.74GSTDD213 pKa = 3.55FFGDD217 pKa = 3.56YY218 pKa = 10.57RR219 pKa = 11.84PDD221 pKa = 3.18QHH223 pKa = 6.04EE224 pKa = 4.36TLVVDD229 pKa = 5.11DD230 pKa = 6.51FYY232 pKa = 11.94SNWKK236 pKa = 8.04YY237 pKa = 6.84TTFLQVCDD245 pKa = 4.41RR246 pKa = 11.84YY247 pKa = 8.69PTEE250 pKa = 3.81VHH252 pKa = 5.73TKK254 pKa = 9.97GGFRR258 pKa = 11.84QLLVRR263 pKa = 11.84HH264 pKa = 6.0IVFTSNHH271 pKa = 5.91SPGVWYY277 pKa = 9.29PNVLADD283 pKa = 3.61ADD285 pKa = 3.91RR286 pKa = 11.84RR287 pKa = 11.84EE288 pKa = 4.18SFNRR292 pKa = 11.84RR293 pKa = 11.84IHH295 pKa = 5.9NIIWFTPDD303 pKa = 3.27GYY305 pKa = 9.39WVKK308 pKa = 10.65KK309 pKa = 8.81GTLPWPCPFLQPLAWNHH326 pKa = 4.4VLRR329 pKa = 11.84NEE331 pKa = 4.11MLPIALLPQNQNLTIHH347 pKa = 6.18EE348 pKa = 4.46RR349 pKa = 11.84GILDD353 pKa = 4.06PSQLFVV359 pKa = 3.4
Molecular weight: 41.71 kDa
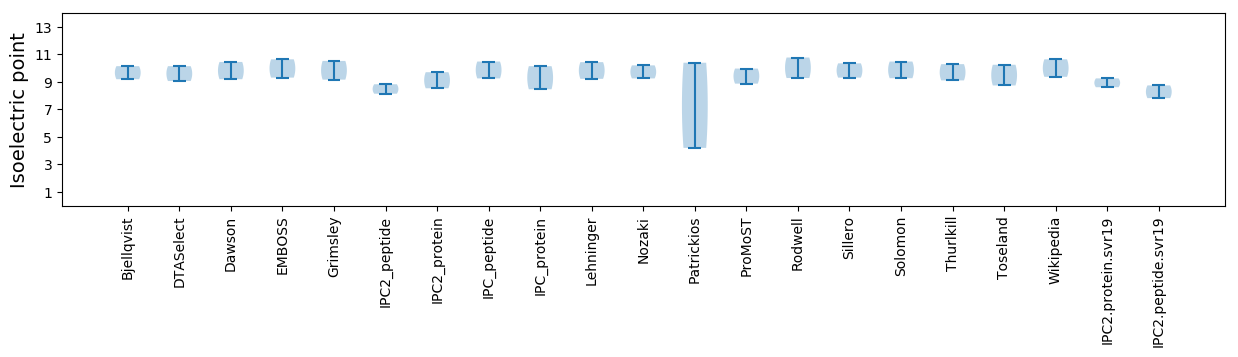
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345BKB0|A0A345BKB0_9VIRU Coat protein OS=Holcus lanatus-associated virus OX=2282643 PE=4 SV=1
MM1 pKa = 7.68GPMRR5 pKa = 11.84QYY7 pKa = 11.32KK8 pKa = 8.33PTGSPRR14 pKa = 11.84AGQRR18 pKa = 11.84AYY20 pKa = 9.92KK21 pKa = 9.82SVYY24 pKa = 8.6SRR26 pKa = 11.84RR27 pKa = 11.84PNRR30 pKa = 11.84PMMGPSNKK38 pKa = 9.51AVSLLKK44 pKa = 9.89MGYY47 pKa = 9.88LNRR50 pKa = 11.84RR51 pKa = 11.84GQLNRR56 pKa = 11.84EE57 pKa = 3.76TGYY60 pKa = 11.32VDD62 pKa = 4.96LASQVFNGNTTGTIVLAATIVQGAAVTQRR91 pKa = 11.84VGKK94 pKa = 9.76KK95 pKa = 8.99VLWKK99 pKa = 10.18GMQIRR104 pKa = 11.84GYY106 pKa = 10.19AQADD110 pKa = 3.44NTTLVTGGTWILVYY124 pKa = 10.4DD125 pKa = 4.21KK126 pKa = 10.6RR127 pKa = 11.84PTGALPAITDD137 pKa = 3.37ILVSVNSSAFLNDD150 pKa = 3.34ANSGRR155 pKa = 11.84FKK157 pKa = 10.58ILARR161 pKa = 11.84RR162 pKa = 11.84SYY164 pKa = 11.39SLTGNVATAGQQNDD178 pKa = 3.7STFQMIDD185 pKa = 3.24EE186 pKa = 4.23YY187 pKa = 11.79VKK189 pKa = 10.94LKK191 pKa = 10.58NLPSVFKK198 pKa = 10.79AVGSGDD204 pKa = 3.08IGDD207 pKa = 4.04IEE209 pKa = 4.47EE210 pKa = 4.36GALYY214 pKa = 10.39LVTVGTTPTGTADD227 pKa = 4.53MIFQLAARR235 pKa = 11.84TRR237 pKa = 11.84FLDD240 pKa = 3.39VV241 pKa = 3.29
MM1 pKa = 7.68GPMRR5 pKa = 11.84QYY7 pKa = 11.32KK8 pKa = 8.33PTGSPRR14 pKa = 11.84AGQRR18 pKa = 11.84AYY20 pKa = 9.92KK21 pKa = 9.82SVYY24 pKa = 8.6SRR26 pKa = 11.84RR27 pKa = 11.84PNRR30 pKa = 11.84PMMGPSNKK38 pKa = 9.51AVSLLKK44 pKa = 9.89MGYY47 pKa = 9.88LNRR50 pKa = 11.84RR51 pKa = 11.84GQLNRR56 pKa = 11.84EE57 pKa = 3.76TGYY60 pKa = 11.32VDD62 pKa = 4.96LASQVFNGNTTGTIVLAATIVQGAAVTQRR91 pKa = 11.84VGKK94 pKa = 9.76KK95 pKa = 8.99VLWKK99 pKa = 10.18GMQIRR104 pKa = 11.84GYY106 pKa = 10.19AQADD110 pKa = 3.44NTTLVTGGTWILVYY124 pKa = 10.4DD125 pKa = 4.21KK126 pKa = 10.6RR127 pKa = 11.84PTGALPAITDD137 pKa = 3.37ILVSVNSSAFLNDD150 pKa = 3.34ANSGRR155 pKa = 11.84FKK157 pKa = 10.58ILARR161 pKa = 11.84RR162 pKa = 11.84SYY164 pKa = 11.39SLTGNVATAGQQNDD178 pKa = 3.7STFQMIDD185 pKa = 3.24EE186 pKa = 4.23YY187 pKa = 11.79VKK189 pKa = 10.94LKK191 pKa = 10.58NLPSVFKK198 pKa = 10.79AVGSGDD204 pKa = 3.08IGDD207 pKa = 4.04IEE209 pKa = 4.47EE210 pKa = 4.36GALYY214 pKa = 10.39LVTVGTTPTGTADD227 pKa = 4.53MIFQLAARR235 pKa = 11.84TRR237 pKa = 11.84FLDD240 pKa = 3.39VV241 pKa = 3.29
Molecular weight: 26.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
600 |
241 |
359 |
300.0 |
33.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.167 ± 1.235 | 1.333 ± 0.839 |
5.0 ± 0.274 | 2.667 ± 0.634 |
4.0 ± 0.69 | 7.5 ± 1.809 |
2.667 ± 1.679 | 4.5 ± 0.04 |
5.0 ± 0.013 | 7.667 ± 0.398 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.333 ± 0.621 | 5.333 ± 0.038 |
6.5 ± 1.48 | 5.333 ± 0.223 |
6.833 ± 0.139 | 5.167 ± 0.666 |
7.333 ± 1.13 | 6.833 ± 0.923 |
2.0 ± 0.737 | 4.833 ± 0.431 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
