
Flavobacterium terrae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
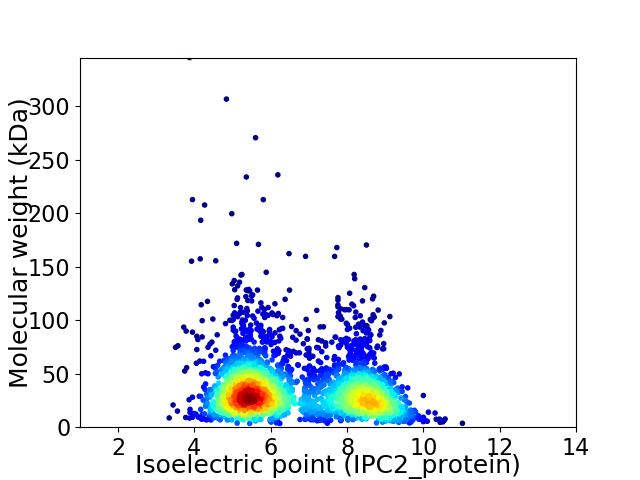
Virtual 2D-PAGE plot for 2883 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6FQ51|A0A1M6FQ51_9FLAO Lipopolysaccharide export system ATP-binding protein OS=Flavobacterium terrae OX=415425 GN=SAMN05444363_2283 PE=4 SV=1
MM1 pKa = 6.41KK2 pKa = 10.16QKK4 pKa = 10.8YY5 pKa = 9.02ILFIIFSLLVFKK17 pKa = 11.06GKK19 pKa = 9.92AQNNYY24 pKa = 8.9SVAPIPFQPFIGSLTPLNALDD45 pKa = 5.14DD46 pKa = 4.29MYY48 pKa = 11.78SNIIPLPFSFDD59 pKa = 4.04FYY61 pKa = 11.46GVTYY65 pKa = 10.67NQMIVSTNGFINFTTTSAGGVSPWSFSQQIPNASFPVTNSILGCFEE111 pKa = 5.08DD112 pKa = 5.18LYY114 pKa = 11.03NNSGTGSITYY124 pKa = 9.47GVYY127 pKa = 9.2GTAPYY132 pKa = 10.43RR133 pKa = 11.84KK134 pKa = 8.52FVVYY138 pKa = 10.45FNDD141 pKa = 3.63QPHH144 pKa = 6.42FSCNTIAISSFQMILSEE161 pKa = 4.14TANTIDD167 pKa = 3.15VQLIEE172 pKa = 4.5RR173 pKa = 11.84QPCLGWNSGRR183 pKa = 11.84GVMGIVKK190 pKa = 10.46DD191 pKa = 3.87GANALAAPGRR201 pKa = 11.84NTGSWTANNEE211 pKa = 3.05AWRR214 pKa = 11.84FYY216 pKa = 10.77RR217 pKa = 11.84AGYY220 pKa = 8.34YY221 pKa = 9.83PNYY224 pKa = 10.6SFVRR228 pKa = 11.84CDD230 pKa = 4.34DD231 pKa = 3.86DD232 pKa = 3.68TDD234 pKa = 4.36GYY236 pKa = 11.0QVFNLSVAANDD247 pKa = 4.05LVPANPSSVIFYY259 pKa = 8.42STLTDD264 pKa = 3.4AQADD268 pKa = 4.16TNSITNQTAYY278 pKa = 9.62TNNSNPQIIYY288 pKa = 10.11AKK290 pKa = 9.74EE291 pKa = 3.45NGIIKK296 pKa = 9.01TVTLSVFDD304 pKa = 4.97CNVDD308 pKa = 3.22ADD310 pKa = 4.05NDD312 pKa = 4.34GVSSSTEE319 pKa = 3.71DD320 pKa = 3.37VNNDD324 pKa = 2.91TNLANDD330 pKa = 3.81DD331 pKa = 4.09TDD333 pKa = 4.78FDD335 pKa = 5.31GIPNYY340 pKa = 10.52LDD342 pKa = 4.06NDD344 pKa = 4.18DD345 pKa = 6.02DD346 pKa = 5.91GDD348 pKa = 4.13LVLTNIEE355 pKa = 4.27YY356 pKa = 10.91VFGKK360 pKa = 10.09NVSALLDD367 pKa = 3.7TDD369 pKa = 3.61NDD371 pKa = 4.91GIPNYY376 pKa = 10.35LDD378 pKa = 4.14NDD380 pKa = 4.21DD381 pKa = 5.78DD382 pKa = 6.08GDD384 pKa = 3.99GSLTFLEE391 pKa = 5.53DD392 pKa = 3.51YY393 pKa = 10.9NHH395 pKa = 7.61DD396 pKa = 4.1GNPVNDD402 pKa = 3.65DD403 pKa = 3.78TNFNNIPDD411 pKa = 4.24YY412 pKa = 11.17LDD414 pKa = 3.2QAVTLGVSQVRR425 pKa = 11.84IDD427 pKa = 3.92DD428 pKa = 4.09NAILMYY434 pKa = 9.7PNPATSMLNILNKK447 pKa = 7.52TTEE450 pKa = 4.03NVMLIEE456 pKa = 5.04IYY458 pKa = 10.61NSNGSRR464 pKa = 11.84AKK466 pKa = 10.46SINSTQSLTTIPVSDD481 pKa = 4.34LQSGVYY487 pKa = 8.29FVKK490 pKa = 10.77VIMNNQVVNYY500 pKa = 10.15KK501 pKa = 10.28FIKK504 pKa = 10.38DD505 pKa = 3.28
MM1 pKa = 6.41KK2 pKa = 10.16QKK4 pKa = 10.8YY5 pKa = 9.02ILFIIFSLLVFKK17 pKa = 11.06GKK19 pKa = 9.92AQNNYY24 pKa = 8.9SVAPIPFQPFIGSLTPLNALDD45 pKa = 5.14DD46 pKa = 4.29MYY48 pKa = 11.78SNIIPLPFSFDD59 pKa = 4.04FYY61 pKa = 11.46GVTYY65 pKa = 10.67NQMIVSTNGFINFTTTSAGGVSPWSFSQQIPNASFPVTNSILGCFEE111 pKa = 5.08DD112 pKa = 5.18LYY114 pKa = 11.03NNSGTGSITYY124 pKa = 9.47GVYY127 pKa = 9.2GTAPYY132 pKa = 10.43RR133 pKa = 11.84KK134 pKa = 8.52FVVYY138 pKa = 10.45FNDD141 pKa = 3.63QPHH144 pKa = 6.42FSCNTIAISSFQMILSEE161 pKa = 4.14TANTIDD167 pKa = 3.15VQLIEE172 pKa = 4.5RR173 pKa = 11.84QPCLGWNSGRR183 pKa = 11.84GVMGIVKK190 pKa = 10.46DD191 pKa = 3.87GANALAAPGRR201 pKa = 11.84NTGSWTANNEE211 pKa = 3.05AWRR214 pKa = 11.84FYY216 pKa = 10.77RR217 pKa = 11.84AGYY220 pKa = 8.34YY221 pKa = 9.83PNYY224 pKa = 10.6SFVRR228 pKa = 11.84CDD230 pKa = 4.34DD231 pKa = 3.86DD232 pKa = 3.68TDD234 pKa = 4.36GYY236 pKa = 11.0QVFNLSVAANDD247 pKa = 4.05LVPANPSSVIFYY259 pKa = 8.42STLTDD264 pKa = 3.4AQADD268 pKa = 4.16TNSITNQTAYY278 pKa = 9.62TNNSNPQIIYY288 pKa = 10.11AKK290 pKa = 9.74EE291 pKa = 3.45NGIIKK296 pKa = 9.01TVTLSVFDD304 pKa = 4.97CNVDD308 pKa = 3.22ADD310 pKa = 4.05NDD312 pKa = 4.34GVSSSTEE319 pKa = 3.71DD320 pKa = 3.37VNNDD324 pKa = 2.91TNLANDD330 pKa = 3.81DD331 pKa = 4.09TDD333 pKa = 4.78FDD335 pKa = 5.31GIPNYY340 pKa = 10.52LDD342 pKa = 4.06NDD344 pKa = 4.18DD345 pKa = 6.02DD346 pKa = 5.91GDD348 pKa = 4.13LVLTNIEE355 pKa = 4.27YY356 pKa = 10.91VFGKK360 pKa = 10.09NVSALLDD367 pKa = 3.7TDD369 pKa = 3.61NDD371 pKa = 4.91GIPNYY376 pKa = 10.35LDD378 pKa = 4.14NDD380 pKa = 4.21DD381 pKa = 5.78DD382 pKa = 6.08GDD384 pKa = 3.99GSLTFLEE391 pKa = 5.53DD392 pKa = 3.51YY393 pKa = 10.9NHH395 pKa = 7.61DD396 pKa = 4.1GNPVNDD402 pKa = 3.65DD403 pKa = 3.78TNFNNIPDD411 pKa = 4.24YY412 pKa = 11.17LDD414 pKa = 3.2QAVTLGVSQVRR425 pKa = 11.84IDD427 pKa = 3.92DD428 pKa = 4.09NAILMYY434 pKa = 9.7PNPATSMLNILNKK447 pKa = 7.52TTEE450 pKa = 4.03NVMLIEE456 pKa = 5.04IYY458 pKa = 10.61NSNGSRR464 pKa = 11.84AKK466 pKa = 10.46SINSTQSLTTIPVSDD481 pKa = 4.34LQSGVYY487 pKa = 8.29FVKK490 pKa = 10.77VIMNNQVVNYY500 pKa = 10.15KK501 pKa = 10.28FIKK504 pKa = 10.38DD505 pKa = 3.28
Molecular weight: 55.65 kDa
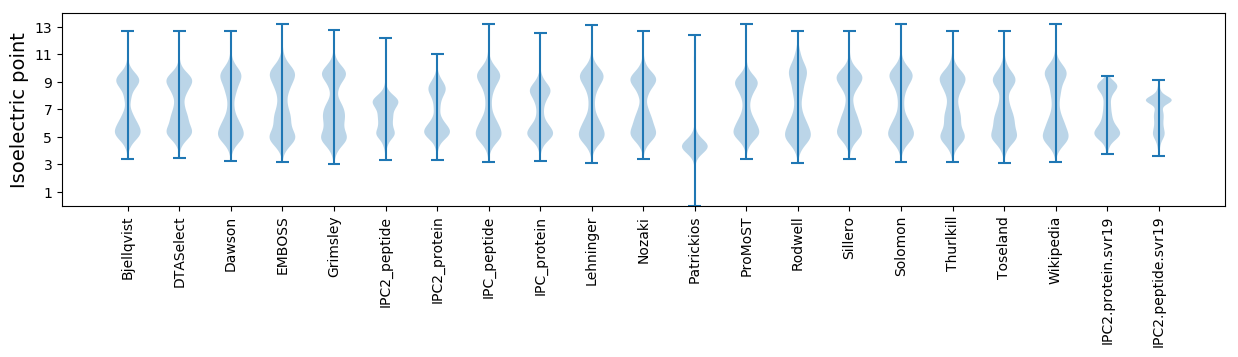
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6BEJ7|A0A1M6BEJ7_9FLAO NADH-quinone oxidoreductase subunit N OS=Flavobacterium terrae OX=415425 GN=nuoN PE=3 SV=1
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.35KK16 pKa = 10.31RR17 pKa = 11.84KK18 pKa = 9.82KK19 pKa = 8.65RR20 pKa = 11.84ARR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.91KK27 pKa = 10.59KK28 pKa = 9.82KK29 pKa = 10.51KK30 pKa = 10.04
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.35KK16 pKa = 10.31RR17 pKa = 11.84KK18 pKa = 9.82KK19 pKa = 8.65RR20 pKa = 11.84ARR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.91KK27 pKa = 10.59KK28 pKa = 9.82KK29 pKa = 10.51KK30 pKa = 10.04
Molecular weight: 3.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
969809 |
29 |
3322 |
336.4 |
38.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.065 ± 0.058 | 0.811 ± 0.016 |
5.282 ± 0.035 | 6.572 ± 0.056 |
5.461 ± 0.04 | 6.184 ± 0.049 |
1.644 ± 0.022 | 8.114 ± 0.052 |
8.33 ± 0.069 | 8.996 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.187 ± 0.027 | 6.726 ± 0.053 |
3.282 ± 0.031 | 3.423 ± 0.027 |
3.02 ± 0.029 | 6.706 ± 0.041 |
5.817 ± 0.067 | 6.257 ± 0.038 |
0.977 ± 0.019 | 4.147 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
