
Betapapillomavirus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
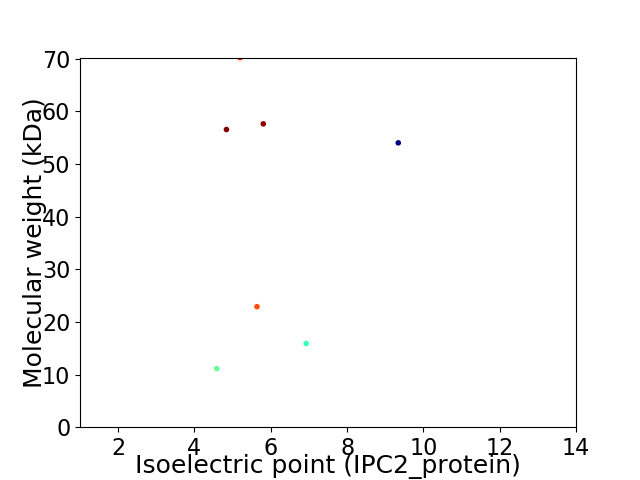
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2AL34|A0A2D2AL34_9PAPI Regulatory protein E2 OS=Betapapillomavirus 5 OX=334209 GN=E2 PE=3 SV=1
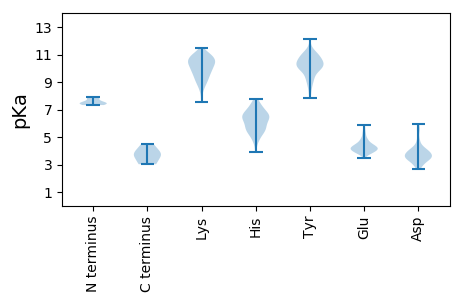
MM1 pKa = 7.43IGKK4 pKa = 8.97EE5 pKa = 3.79VTIPDD10 pKa = 4.17IEE12 pKa = 5.64LEE14 pKa = 4.19LQDD17 pKa = 4.42LVQPIDD23 pKa = 3.48LHH25 pKa = 7.78CDD27 pKa = 2.97EE28 pKa = 5.09VLPEE32 pKa = 3.94EE33 pKa = 4.52SEE35 pKa = 4.24NLSEE39 pKa = 4.18SSQAEE44 pKa = 4.21VEE46 pKa = 4.15PEE48 pKa = 3.96RR49 pKa = 11.84ILFKK53 pKa = 10.62IVAPCGGCEE62 pKa = 3.8TRR64 pKa = 11.84LKK66 pKa = 10.39IHH68 pKa = 6.63IASTRR73 pKa = 11.84FGIRR77 pKa = 11.84CLEE80 pKa = 3.87EE81 pKa = 4.56LLLSEE86 pKa = 5.47LCLLCPVCRR95 pKa = 11.84NGRR98 pKa = 11.84QQ99 pKa = 3.02
MM1 pKa = 7.43IGKK4 pKa = 8.97EE5 pKa = 3.79VTIPDD10 pKa = 4.17IEE12 pKa = 5.64LEE14 pKa = 4.19LQDD17 pKa = 4.42LVQPIDD23 pKa = 3.48LHH25 pKa = 7.78CDD27 pKa = 2.97EE28 pKa = 5.09VLPEE32 pKa = 3.94EE33 pKa = 4.52SEE35 pKa = 4.24NLSEE39 pKa = 4.18SSQAEE44 pKa = 4.21VEE46 pKa = 4.15PEE48 pKa = 3.96RR49 pKa = 11.84ILFKK53 pKa = 10.62IVAPCGGCEE62 pKa = 3.8TRR64 pKa = 11.84LKK66 pKa = 10.39IHH68 pKa = 6.63IASTRR73 pKa = 11.84FGIRR77 pKa = 11.84CLEE80 pKa = 3.87EE81 pKa = 4.56LLLSEE86 pKa = 5.47LCLLCPVCRR95 pKa = 11.84NGRR98 pKa = 11.84QQ99 pKa = 3.02
Molecular weight: 11.12 kDa
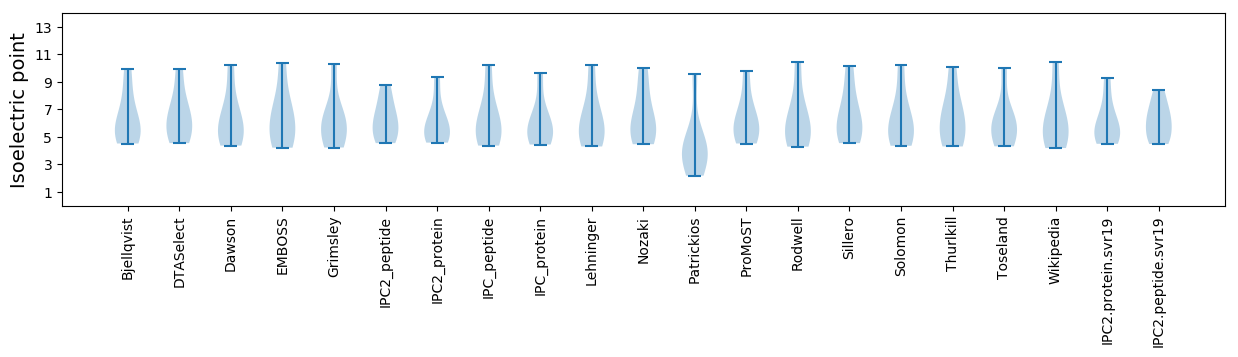
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2AL37|A0A2D2AL37_9PAPI Minor capsid protein L2 OS=Betapapillomavirus 5 OX=334209 GN=L2 PE=3 SV=1
MM1 pKa = 7.45EE2 pKa = 5.84ALNQRR7 pKa = 11.84FNVLQDD13 pKa = 3.4QLMDD17 pKa = 3.31IYY19 pKa = 11.21EE20 pKa = 4.53RR21 pKa = 11.84GSDD24 pKa = 3.43TLEE27 pKa = 3.75EE28 pKa = 4.33QIKK31 pKa = 9.81HH32 pKa = 3.94WTLLKK37 pKa = 10.1QEE39 pKa = 4.31QIILHH44 pKa = 4.89YY45 pKa = 10.14ARR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84GIMRR53 pKa = 11.84LGYY56 pKa = 10.55HH57 pKa = 6.55PVPTLAVSEE66 pKa = 4.87LKK68 pKa = 10.95AKK70 pKa = 10.45DD71 pKa = 3.73AIAMVLHH78 pKa = 6.57LEE80 pKa = 4.13NLKK83 pKa = 10.32QSPYY87 pKa = 10.46KK88 pKa = 10.42DD89 pKa = 3.96EE90 pKa = 4.36PWTLINTSLEE100 pKa = 4.4TFRR103 pKa = 11.84SPPANCFKK111 pKa = 10.83KK112 pKa = 10.47GPQNVEE118 pKa = 3.83VLFDD122 pKa = 4.19GDD124 pKa = 4.09PEE126 pKa = 4.33NVMLYY131 pKa = 7.93TAWKK135 pKa = 8.95FIYY138 pKa = 10.86YY139 pKa = 10.09EE140 pKa = 4.13DD141 pKa = 5.56TEE143 pKa = 4.56GQWQKK148 pKa = 9.84TEE150 pKa = 3.79GHH152 pKa = 6.43IDD154 pKa = 3.57YY155 pKa = 10.98AGLYY159 pKa = 8.59YY160 pKa = 10.57VEE162 pKa = 5.6GGIKK166 pKa = 9.76HH167 pKa = 6.0YY168 pKa = 10.57YY169 pKa = 10.02VEE171 pKa = 4.78FNVDD175 pKa = 2.93AQRR178 pKa = 11.84FGTRR182 pKa = 11.84GEE184 pKa = 3.95WEE186 pKa = 3.9VRR188 pKa = 11.84FNGEE192 pKa = 3.88TLFAPVTSSSPSPLEE207 pKa = 4.35EE208 pKa = 4.55IGEE211 pKa = 4.4RR212 pKa = 11.84PKK214 pKa = 11.31SPDD217 pKa = 3.27VPEE220 pKa = 4.3TTTDD224 pKa = 2.82TGTLQRR230 pKa = 11.84EE231 pKa = 4.27TRR233 pKa = 11.84HH234 pKa = 4.91TEE236 pKa = 3.74RR237 pKa = 11.84AEE239 pKa = 3.89STLPKK244 pKa = 9.62GRR246 pKa = 11.84RR247 pKa = 11.84YY248 pKa = 8.84EE249 pKa = 4.16RR250 pKa = 11.84KK251 pKa = 9.12EE252 pKa = 3.77SSPTSTSLRR261 pKa = 11.84GRR263 pKa = 11.84QKK265 pKa = 10.9VSGHH269 pKa = 4.82TSRR272 pKa = 11.84KK273 pKa = 8.43KK274 pKa = 8.75EE275 pKa = 3.9SRR277 pKa = 11.84RR278 pKa = 11.84SRR280 pKa = 11.84STSSGRR286 pKa = 11.84LGTDD290 pKa = 2.8QAAARR295 pKa = 11.84GRR297 pKa = 11.84GQRR300 pKa = 11.84QARR303 pKa = 11.84PRR305 pKa = 11.84TRR307 pKa = 11.84SRR309 pKa = 11.84SRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84SRR315 pKa = 11.84SRR317 pKa = 11.84SKK319 pKa = 11.05GGGRR323 pKa = 11.84TTAEE327 pKa = 4.1SRR329 pKa = 11.84RR330 pKa = 11.84GRR332 pKa = 11.84CGGRR336 pKa = 11.84GYY338 pKa = 8.69FTRR341 pKa = 11.84SRR343 pKa = 11.84SRR345 pKa = 11.84SAEE348 pKa = 3.85SRR350 pKa = 11.84STDD353 pKa = 2.89QCGILPSQVGTGVQSVGRR371 pKa = 11.84KK372 pKa = 7.53YY373 pKa = 10.16SSRR376 pKa = 11.84VEE378 pKa = 3.96RR379 pKa = 11.84LLAEE383 pKa = 4.47AVDD386 pKa = 4.02PPVILIRR393 pKa = 11.84GPPNTLKK400 pKa = 10.6CYY402 pKa = 10.23RR403 pKa = 11.84YY404 pKa = 9.67RR405 pKa = 11.84EE406 pKa = 4.05KK407 pKa = 11.01EE408 pKa = 4.02RR409 pKa = 11.84LKK411 pKa = 11.04GFYY414 pKa = 10.43DD415 pKa = 3.69RR416 pKa = 11.84FSTTWSWVGSEE427 pKa = 3.91GNARR431 pKa = 11.84LGRR434 pKa = 11.84ARR436 pKa = 11.84MLICFLNEE444 pKa = 3.55EE445 pKa = 3.96QRR447 pKa = 11.84EE448 pKa = 4.05RR449 pKa = 11.84FIDD452 pKa = 4.05KK453 pKa = 9.45MRR455 pKa = 11.84LPKK458 pKa = 10.68GVDD461 pKa = 2.66WSYY464 pKa = 12.12GNFASII470 pKa = 4.49
MM1 pKa = 7.45EE2 pKa = 5.84ALNQRR7 pKa = 11.84FNVLQDD13 pKa = 3.4QLMDD17 pKa = 3.31IYY19 pKa = 11.21EE20 pKa = 4.53RR21 pKa = 11.84GSDD24 pKa = 3.43TLEE27 pKa = 3.75EE28 pKa = 4.33QIKK31 pKa = 9.81HH32 pKa = 3.94WTLLKK37 pKa = 10.1QEE39 pKa = 4.31QIILHH44 pKa = 4.89YY45 pKa = 10.14ARR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84GIMRR53 pKa = 11.84LGYY56 pKa = 10.55HH57 pKa = 6.55PVPTLAVSEE66 pKa = 4.87LKK68 pKa = 10.95AKK70 pKa = 10.45DD71 pKa = 3.73AIAMVLHH78 pKa = 6.57LEE80 pKa = 4.13NLKK83 pKa = 10.32QSPYY87 pKa = 10.46KK88 pKa = 10.42DD89 pKa = 3.96EE90 pKa = 4.36PWTLINTSLEE100 pKa = 4.4TFRR103 pKa = 11.84SPPANCFKK111 pKa = 10.83KK112 pKa = 10.47GPQNVEE118 pKa = 3.83VLFDD122 pKa = 4.19GDD124 pKa = 4.09PEE126 pKa = 4.33NVMLYY131 pKa = 7.93TAWKK135 pKa = 8.95FIYY138 pKa = 10.86YY139 pKa = 10.09EE140 pKa = 4.13DD141 pKa = 5.56TEE143 pKa = 4.56GQWQKK148 pKa = 9.84TEE150 pKa = 3.79GHH152 pKa = 6.43IDD154 pKa = 3.57YY155 pKa = 10.98AGLYY159 pKa = 8.59YY160 pKa = 10.57VEE162 pKa = 5.6GGIKK166 pKa = 9.76HH167 pKa = 6.0YY168 pKa = 10.57YY169 pKa = 10.02VEE171 pKa = 4.78FNVDD175 pKa = 2.93AQRR178 pKa = 11.84FGTRR182 pKa = 11.84GEE184 pKa = 3.95WEE186 pKa = 3.9VRR188 pKa = 11.84FNGEE192 pKa = 3.88TLFAPVTSSSPSPLEE207 pKa = 4.35EE208 pKa = 4.55IGEE211 pKa = 4.4RR212 pKa = 11.84PKK214 pKa = 11.31SPDD217 pKa = 3.27VPEE220 pKa = 4.3TTTDD224 pKa = 2.82TGTLQRR230 pKa = 11.84EE231 pKa = 4.27TRR233 pKa = 11.84HH234 pKa = 4.91TEE236 pKa = 3.74RR237 pKa = 11.84AEE239 pKa = 3.89STLPKK244 pKa = 9.62GRR246 pKa = 11.84RR247 pKa = 11.84YY248 pKa = 8.84EE249 pKa = 4.16RR250 pKa = 11.84KK251 pKa = 9.12EE252 pKa = 3.77SSPTSTSLRR261 pKa = 11.84GRR263 pKa = 11.84QKK265 pKa = 10.9VSGHH269 pKa = 4.82TSRR272 pKa = 11.84KK273 pKa = 8.43KK274 pKa = 8.75EE275 pKa = 3.9SRR277 pKa = 11.84RR278 pKa = 11.84SRR280 pKa = 11.84STSSGRR286 pKa = 11.84LGTDD290 pKa = 2.8QAAARR295 pKa = 11.84GRR297 pKa = 11.84GQRR300 pKa = 11.84QARR303 pKa = 11.84PRR305 pKa = 11.84TRR307 pKa = 11.84SRR309 pKa = 11.84SRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84SRR315 pKa = 11.84SRR317 pKa = 11.84SKK319 pKa = 11.05GGGRR323 pKa = 11.84TTAEE327 pKa = 4.1SRR329 pKa = 11.84RR330 pKa = 11.84GRR332 pKa = 11.84CGGRR336 pKa = 11.84GYY338 pKa = 8.69FTRR341 pKa = 11.84SRR343 pKa = 11.84SRR345 pKa = 11.84SAEE348 pKa = 3.85SRR350 pKa = 11.84STDD353 pKa = 2.89QCGILPSQVGTGVQSVGRR371 pKa = 11.84KK372 pKa = 7.53YY373 pKa = 10.16SSRR376 pKa = 11.84VEE378 pKa = 3.96RR379 pKa = 11.84LLAEE383 pKa = 4.47AVDD386 pKa = 4.02PPVILIRR393 pKa = 11.84GPPNTLKK400 pKa = 10.6CYY402 pKa = 10.23RR403 pKa = 11.84YY404 pKa = 9.67RR405 pKa = 11.84EE406 pKa = 4.05KK407 pKa = 11.01EE408 pKa = 4.02RR409 pKa = 11.84LKK411 pKa = 11.04GFYY414 pKa = 10.43DD415 pKa = 3.69RR416 pKa = 11.84FSTTWSWVGSEE427 pKa = 3.91GNARR431 pKa = 11.84LGRR434 pKa = 11.84ARR436 pKa = 11.84MLICFLNEE444 pKa = 3.55EE445 pKa = 3.96QRR447 pKa = 11.84EE448 pKa = 4.05RR449 pKa = 11.84FIDD452 pKa = 4.05KK453 pKa = 9.45MRR455 pKa = 11.84LPKK458 pKa = 10.68GVDD461 pKa = 2.66WSYY464 pKa = 12.12GNFASII470 pKa = 4.49
Molecular weight: 53.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2553 |
99 |
605 |
364.7 |
41.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.327 ± 0.204 | 2.35 ± 0.791 |
5.64 ± 0.483 | 7.521 ± 0.856 |
3.878 ± 0.424 | 7.09 ± 0.84 |
2.115 ± 0.23 | 4.779 ± 0.664 |
4.896 ± 0.76 | 8.931 ± 0.89 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.528 ± 0.457 | 4.152 ± 0.738 |
6.11 ± 1.166 | 4.505 ± 0.338 |
6.972 ± 1.24 | 7.286 ± 0.546 |
5.993 ± 0.977 | 6.11 ± 0.823 |
1.332 ± 0.366 | 3.486 ± 0.313 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
