
Cryobacterium mesophilum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Cryobacterium
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
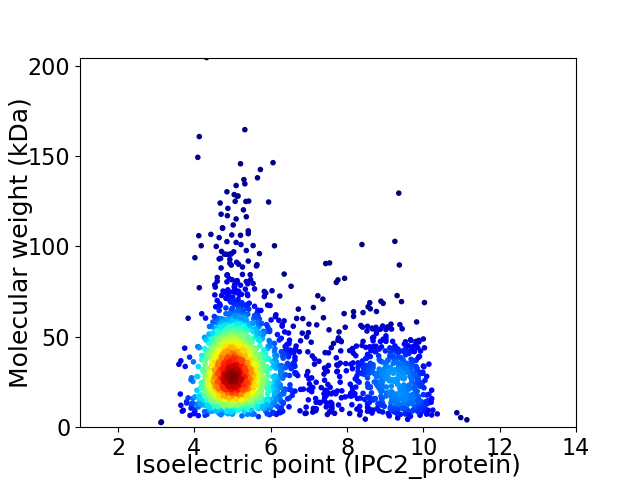
Virtual 2D-PAGE plot for 2302 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
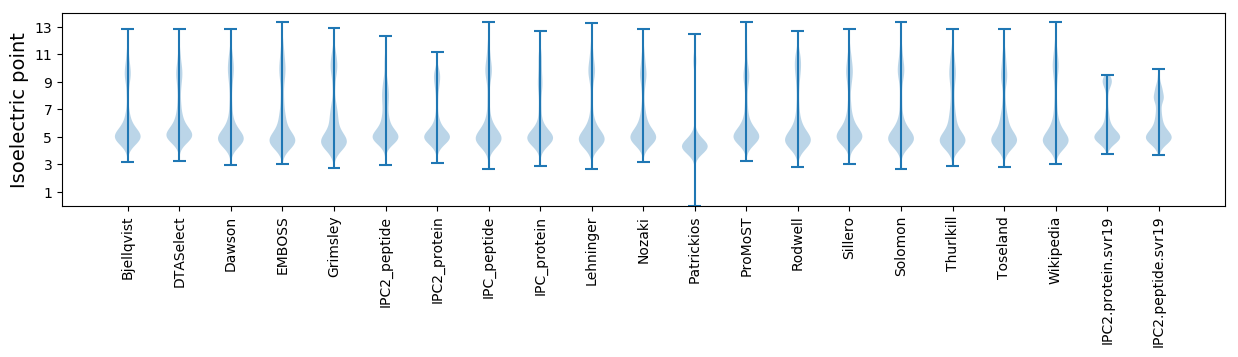
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R8VEJ0|A0A4R8VEJ0_9MICO DNA-binding GntR family transcriptional regulator OS=Cryobacterium mesophilum OX=433647 GN=BKA04_001787 PE=4 SV=1
MM1 pKa = 7.46SISGAPMRR9 pKa = 11.84TSKK12 pKa = 10.58FLSLAAASALLAGGSLLSAGPASASTTVDD41 pKa = 3.03YY42 pKa = 9.2ATFYY46 pKa = 10.36GASDD50 pKa = 3.94FGVEE54 pKa = 4.03SGGYY58 pKa = 5.63PTGVDD63 pKa = 2.96WFFGDD68 pKa = 3.45VSGTEE73 pKa = 4.4GPHH76 pKa = 5.44QFTNTGLVLNDD87 pKa = 3.86PASGDD92 pKa = 3.77VQILNQNVTTPVDD105 pKa = 3.6AGEE108 pKa = 4.25LASVVQNANIFAQNGEE124 pKa = 4.3WFFQLAFFAEE134 pKa = 5.05GTSDD138 pKa = 3.37TGFTTLRR145 pKa = 11.84PATAGSVSTGDD156 pKa = 3.26PWITSQALGSYY167 pKa = 9.94AAGATASFSDD177 pKa = 4.91LADD180 pKa = 3.4ALYY183 pKa = 10.71AGEE186 pKa = 4.64APQLLAYY193 pKa = 10.18GFFVAAGQTTTINGISWGTQGSSFGLPPTRR223 pKa = 11.84TISPNPITQDD233 pKa = 3.08AFSTAGQGLTLSGTNWFPGVDD254 pKa = 3.39AYY256 pKa = 10.26IFIVDD261 pKa = 3.84HH262 pKa = 6.59NGTTVFEE269 pKa = 4.44DD270 pKa = 3.63TSSFIVDD277 pKa = 3.3SAGNVSVSVVLPSKK291 pKa = 10.36PDD293 pKa = 3.0VGTYY297 pKa = 10.38YY298 pKa = 9.63VTFDD302 pKa = 4.2DD303 pKa = 5.28DD304 pKa = 3.56SFYY307 pKa = 11.51YY308 pKa = 8.7HH309 pKa = 6.79TGVLDD314 pKa = 4.25HH315 pKa = 6.5YY316 pKa = 11.09VSAPGGGEE324 pKa = 4.17GEE326 pKa = 4.34GSNAQGIQISVIAAAPEE343 pKa = 3.85LAATGAEE350 pKa = 4.22SGTLIAGGILLLILGGVVLIGSRR373 pKa = 11.84RR374 pKa = 11.84RR375 pKa = 11.84RR376 pKa = 11.84AQLGG380 pKa = 3.42
MM1 pKa = 7.46SISGAPMRR9 pKa = 11.84TSKK12 pKa = 10.58FLSLAAASALLAGGSLLSAGPASASTTVDD41 pKa = 3.03YY42 pKa = 9.2ATFYY46 pKa = 10.36GASDD50 pKa = 3.94FGVEE54 pKa = 4.03SGGYY58 pKa = 5.63PTGVDD63 pKa = 2.96WFFGDD68 pKa = 3.45VSGTEE73 pKa = 4.4GPHH76 pKa = 5.44QFTNTGLVLNDD87 pKa = 3.86PASGDD92 pKa = 3.77VQILNQNVTTPVDD105 pKa = 3.6AGEE108 pKa = 4.25LASVVQNANIFAQNGEE124 pKa = 4.3WFFQLAFFAEE134 pKa = 5.05GTSDD138 pKa = 3.37TGFTTLRR145 pKa = 11.84PATAGSVSTGDD156 pKa = 3.26PWITSQALGSYY167 pKa = 9.94AAGATASFSDD177 pKa = 4.91LADD180 pKa = 3.4ALYY183 pKa = 10.71AGEE186 pKa = 4.64APQLLAYY193 pKa = 10.18GFFVAAGQTTTINGISWGTQGSSFGLPPTRR223 pKa = 11.84TISPNPITQDD233 pKa = 3.08AFSTAGQGLTLSGTNWFPGVDD254 pKa = 3.39AYY256 pKa = 10.26IFIVDD261 pKa = 3.84HH262 pKa = 6.59NGTTVFEE269 pKa = 4.44DD270 pKa = 3.63TSSFIVDD277 pKa = 3.3SAGNVSVSVVLPSKK291 pKa = 10.36PDD293 pKa = 3.0VGTYY297 pKa = 10.38YY298 pKa = 9.63VTFDD302 pKa = 4.2DD303 pKa = 5.28DD304 pKa = 3.56SFYY307 pKa = 11.51YY308 pKa = 8.7HH309 pKa = 6.79TGVLDD314 pKa = 4.25HH315 pKa = 6.5YY316 pKa = 11.09VSAPGGGEE324 pKa = 4.17GEE326 pKa = 4.34GSNAQGIQISVIAAAPEE343 pKa = 3.85LAATGAEE350 pKa = 4.22SGTLIAGGILLLILGGVVLIGSRR373 pKa = 11.84RR374 pKa = 11.84RR375 pKa = 11.84RR376 pKa = 11.84AQLGG380 pKa = 3.42
Molecular weight: 38.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V3I9Q4|A0A4V3I9Q4_9MICO Ribonucleoside-diphosphate reductase OS=Cryobacterium mesophilum OX=433647 GN=BKA04_001889 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
748348 |
22 |
1971 |
325.1 |
34.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.628 ± 0.069 | 0.511 ± 0.013 |
6.103 ± 0.042 | 5.511 ± 0.046 |
3.316 ± 0.026 | 8.932 ± 0.04 |
1.963 ± 0.026 | 5.107 ± 0.039 |
2.441 ± 0.038 | 10.253 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.859 ± 0.019 | 2.268 ± 0.027 |
5.192 ± 0.036 | 2.716 ± 0.026 |
6.849 ± 0.049 | 6.127 ± 0.038 |
5.969 ± 0.038 | 8.787 ± 0.049 |
1.444 ± 0.018 | 2.025 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
