
Marinobacter similis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Marinobacter
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
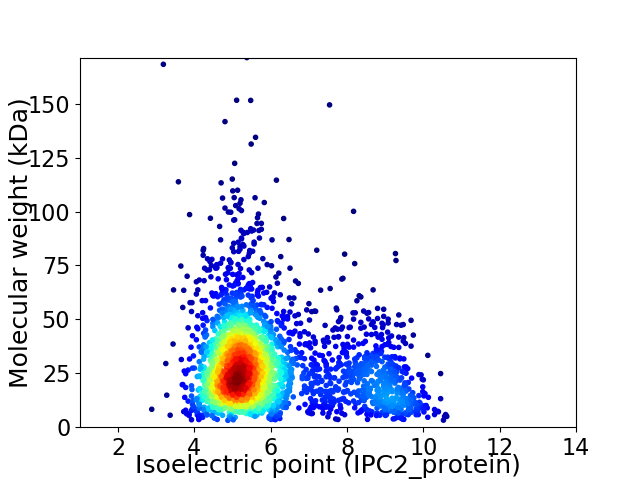
Virtual 2D-PAGE plot for 2692 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5YMT3|W5YMT3_9ALTE LysR family transcriptional regulator OS=Marinobacter similis OX=1420916 GN=AU14_01415 PE=3 SV=1
MM1 pKa = 7.43SVFKK5 pKa = 10.97ALGLGSALILLGGCQSWRR23 pKa = 11.84FQTVEE28 pKa = 4.25EE29 pKa = 4.71LPPPAAEE36 pKa = 4.9PEE38 pKa = 4.24TSQPGEE44 pKa = 4.13VEE46 pKa = 3.65VRR48 pKa = 11.84YY49 pKa = 10.23YY50 pKa = 11.49DD51 pKa = 3.7NLDD54 pKa = 3.46GEE56 pKa = 5.37EE57 pKa = 4.43IDD59 pKa = 5.0TMTTSAKK66 pKa = 10.22FPSNPDD72 pKa = 2.85SVASLNSLEE81 pKa = 4.85DD82 pKa = 3.66PEE84 pKa = 5.45NRR86 pKa = 11.84AEE88 pKa = 4.62DD89 pKa = 3.51YY90 pKa = 10.95GSYY93 pKa = 10.72VRR95 pKa = 11.84GFIKK99 pKa = 10.52PPTSGEE105 pKa = 3.71YY106 pKa = 10.44RR107 pKa = 11.84FFISGNDD114 pKa = 3.28QAEE117 pKa = 4.56FWLSTSDD124 pKa = 3.57SPEE127 pKa = 4.1NVDD130 pKa = 5.11LMAMTPAWTSINEE143 pKa = 3.72FDD145 pKa = 4.62KK146 pKa = 11.51YY147 pKa = 11.05SSQTSRR153 pKa = 11.84YY154 pKa = 7.45ITLDD158 pKa = 2.95SSKK161 pKa = 10.55RR162 pKa = 11.84YY163 pKa = 9.48YY164 pKa = 10.88FEE166 pKa = 4.5VLQKK170 pKa = 10.79EE171 pKa = 5.04GFGSDD176 pKa = 3.01HH177 pKa = 6.88FAVAWEE183 pKa = 4.74GPGISQEE190 pKa = 4.51VIGSSYY196 pKa = 10.0IYY198 pKa = 10.38SYY200 pKa = 11.4AKK202 pKa = 9.87PAIDD206 pKa = 3.4TGQTGQEE213 pKa = 4.18AYY215 pKa = 10.8NLGYY219 pKa = 10.26RR220 pKa = 11.84VGYY223 pKa = 10.47VDD225 pKa = 3.51GTEE228 pKa = 3.97GLSFNPAFPPADD240 pKa = 3.42QDD242 pKa = 3.67QDD244 pKa = 3.95GIYY247 pKa = 10.68DD248 pKa = 3.26NWEE251 pKa = 4.05VVNGLNPNDD260 pKa = 4.39SNDD263 pKa = 3.55ATSDD267 pKa = 3.64PDD269 pKa = 3.93GDD271 pKa = 4.19LLAAADD277 pKa = 4.07EE278 pKa = 4.38FLIGTSEE285 pKa = 3.93NTADD289 pKa = 4.05SDD291 pKa = 4.51GDD293 pKa = 4.17GIPDD297 pKa = 3.32GVEE300 pKa = 3.91FANEE304 pKa = 3.99LDD306 pKa = 3.71PLYY309 pKa = 10.87KK310 pKa = 9.79QDD312 pKa = 4.13AQEE315 pKa = 5.24DD316 pKa = 4.34LDD318 pKa = 4.0NDD320 pKa = 4.33GYY322 pKa = 11.78SNLEE326 pKa = 3.99EE327 pKa = 4.35YY328 pKa = 10.81VAGTVINDD336 pKa = 3.71PDD338 pKa = 4.49DD339 pKa = 4.88APVAAPSPEE348 pKa = 4.25PEE350 pKa = 4.59PEE352 pKa = 4.22PEE354 pKa = 4.48PEE356 pKa = 4.27PEE358 pKa = 4.09PTPSPTEE365 pKa = 3.79EE366 pKa = 3.81PSYY369 pKa = 10.72VAGFGAQFFEE379 pKa = 4.34GTSFNRR385 pKa = 11.84FVLTDD390 pKa = 2.81VHH392 pKa = 6.14EE393 pKa = 4.61AVNFDD398 pKa = 3.45WGRR401 pKa = 11.84GQPLAEE407 pKa = 4.37LPEE410 pKa = 4.5DD411 pKa = 3.36QFSIRR416 pKa = 11.84WNGIFTAPHH425 pKa = 5.38QSGDD429 pKa = 2.8NDD431 pKa = 3.34YY432 pKa = 11.14RR433 pKa = 11.84FTVSTNDD440 pKa = 3.39GVRR443 pKa = 11.84MFVNGEE449 pKa = 3.98LVIDD453 pKa = 4.55DD454 pKa = 4.0WSEE457 pKa = 4.15HH458 pKa = 5.58PTTTYY463 pKa = 10.24TYY465 pKa = 10.72DD466 pKa = 3.35RR467 pKa = 11.84SLAAGEE473 pKa = 4.32KK474 pKa = 10.27LSLTIEE480 pKa = 4.25YY481 pKa = 10.85FEE483 pKa = 4.59GTGSAVAQYY492 pKa = 10.99SATNLTSAEE501 pKa = 4.1NLSTRR506 pKa = 11.84ATVATPDD513 pKa = 3.72PSTSNTLDD521 pKa = 3.41SDD523 pKa = 3.8NDD525 pKa = 4.73GIPDD529 pKa = 3.32TWEE532 pKa = 3.87LRR534 pKa = 11.84NGTSVWVPDD543 pKa = 3.5GGVVNNSQGTSNVEE557 pKa = 3.74AYY559 pKa = 9.98QAGLTPYY566 pKa = 9.71TLATVEE572 pKa = 4.23TDD574 pKa = 3.56GKK576 pKa = 7.76PTSGSGSDD584 pKa = 3.54TGDD587 pKa = 2.93TTTPDD592 pKa = 3.53EE593 pKa = 4.78PSTTEE598 pKa = 3.64PAPPEE603 pKa = 4.26TGSGSVTLTWTAPLTRR619 pKa = 11.84LDD621 pKa = 3.79GSSIALSEE629 pKa = 4.13IASYY633 pKa = 9.77TIDD636 pKa = 3.4YY637 pKa = 9.62GQDD640 pKa = 3.16VNNLEE645 pKa = 4.33EE646 pKa = 4.36SVSVGGDD653 pKa = 3.09QTSYY657 pKa = 11.61SFDD660 pKa = 3.46NLASGTWYY668 pKa = 10.56FKK670 pKa = 10.88IEE672 pKa = 4.14VVDD675 pKa = 3.95TNGLSSPPSNPVSTQVQQ692 pKa = 3.28
MM1 pKa = 7.43SVFKK5 pKa = 10.97ALGLGSALILLGGCQSWRR23 pKa = 11.84FQTVEE28 pKa = 4.25EE29 pKa = 4.71LPPPAAEE36 pKa = 4.9PEE38 pKa = 4.24TSQPGEE44 pKa = 4.13VEE46 pKa = 3.65VRR48 pKa = 11.84YY49 pKa = 10.23YY50 pKa = 11.49DD51 pKa = 3.7NLDD54 pKa = 3.46GEE56 pKa = 5.37EE57 pKa = 4.43IDD59 pKa = 5.0TMTTSAKK66 pKa = 10.22FPSNPDD72 pKa = 2.85SVASLNSLEE81 pKa = 4.85DD82 pKa = 3.66PEE84 pKa = 5.45NRR86 pKa = 11.84AEE88 pKa = 4.62DD89 pKa = 3.51YY90 pKa = 10.95GSYY93 pKa = 10.72VRR95 pKa = 11.84GFIKK99 pKa = 10.52PPTSGEE105 pKa = 3.71YY106 pKa = 10.44RR107 pKa = 11.84FFISGNDD114 pKa = 3.28QAEE117 pKa = 4.56FWLSTSDD124 pKa = 3.57SPEE127 pKa = 4.1NVDD130 pKa = 5.11LMAMTPAWTSINEE143 pKa = 3.72FDD145 pKa = 4.62KK146 pKa = 11.51YY147 pKa = 11.05SSQTSRR153 pKa = 11.84YY154 pKa = 7.45ITLDD158 pKa = 2.95SSKK161 pKa = 10.55RR162 pKa = 11.84YY163 pKa = 9.48YY164 pKa = 10.88FEE166 pKa = 4.5VLQKK170 pKa = 10.79EE171 pKa = 5.04GFGSDD176 pKa = 3.01HH177 pKa = 6.88FAVAWEE183 pKa = 4.74GPGISQEE190 pKa = 4.51VIGSSYY196 pKa = 10.0IYY198 pKa = 10.38SYY200 pKa = 11.4AKK202 pKa = 9.87PAIDD206 pKa = 3.4TGQTGQEE213 pKa = 4.18AYY215 pKa = 10.8NLGYY219 pKa = 10.26RR220 pKa = 11.84VGYY223 pKa = 10.47VDD225 pKa = 3.51GTEE228 pKa = 3.97GLSFNPAFPPADD240 pKa = 3.42QDD242 pKa = 3.67QDD244 pKa = 3.95GIYY247 pKa = 10.68DD248 pKa = 3.26NWEE251 pKa = 4.05VVNGLNPNDD260 pKa = 4.39SNDD263 pKa = 3.55ATSDD267 pKa = 3.64PDD269 pKa = 3.93GDD271 pKa = 4.19LLAAADD277 pKa = 4.07EE278 pKa = 4.38FLIGTSEE285 pKa = 3.93NTADD289 pKa = 4.05SDD291 pKa = 4.51GDD293 pKa = 4.17GIPDD297 pKa = 3.32GVEE300 pKa = 3.91FANEE304 pKa = 3.99LDD306 pKa = 3.71PLYY309 pKa = 10.87KK310 pKa = 9.79QDD312 pKa = 4.13AQEE315 pKa = 5.24DD316 pKa = 4.34LDD318 pKa = 4.0NDD320 pKa = 4.33GYY322 pKa = 11.78SNLEE326 pKa = 3.99EE327 pKa = 4.35YY328 pKa = 10.81VAGTVINDD336 pKa = 3.71PDD338 pKa = 4.49DD339 pKa = 4.88APVAAPSPEE348 pKa = 4.25PEE350 pKa = 4.59PEE352 pKa = 4.22PEE354 pKa = 4.48PEE356 pKa = 4.27PEE358 pKa = 4.09PTPSPTEE365 pKa = 3.79EE366 pKa = 3.81PSYY369 pKa = 10.72VAGFGAQFFEE379 pKa = 4.34GTSFNRR385 pKa = 11.84FVLTDD390 pKa = 2.81VHH392 pKa = 6.14EE393 pKa = 4.61AVNFDD398 pKa = 3.45WGRR401 pKa = 11.84GQPLAEE407 pKa = 4.37LPEE410 pKa = 4.5DD411 pKa = 3.36QFSIRR416 pKa = 11.84WNGIFTAPHH425 pKa = 5.38QSGDD429 pKa = 2.8NDD431 pKa = 3.34YY432 pKa = 11.14RR433 pKa = 11.84FTVSTNDD440 pKa = 3.39GVRR443 pKa = 11.84MFVNGEE449 pKa = 3.98LVIDD453 pKa = 4.55DD454 pKa = 4.0WSEE457 pKa = 4.15HH458 pKa = 5.58PTTTYY463 pKa = 10.24TYY465 pKa = 10.72DD466 pKa = 3.35RR467 pKa = 11.84SLAAGEE473 pKa = 4.32KK474 pKa = 10.27LSLTIEE480 pKa = 4.25YY481 pKa = 10.85FEE483 pKa = 4.59GTGSAVAQYY492 pKa = 10.99SATNLTSAEE501 pKa = 4.1NLSTRR506 pKa = 11.84ATVATPDD513 pKa = 3.72PSTSNTLDD521 pKa = 3.41SDD523 pKa = 3.8NDD525 pKa = 4.73GIPDD529 pKa = 3.32TWEE532 pKa = 3.87LRR534 pKa = 11.84NGTSVWVPDD543 pKa = 3.5GGVVNNSQGTSNVEE557 pKa = 3.74AYY559 pKa = 9.98QAGLTPYY566 pKa = 9.71TLATVEE572 pKa = 4.23TDD574 pKa = 3.56GKK576 pKa = 7.76PTSGSGSDD584 pKa = 3.54TGDD587 pKa = 2.93TTTPDD592 pKa = 3.53EE593 pKa = 4.78PSTTEE598 pKa = 3.64PAPPEE603 pKa = 4.26TGSGSVTLTWTAPLTRR619 pKa = 11.84LDD621 pKa = 3.79GSSIALSEE629 pKa = 4.13IASYY633 pKa = 9.77TIDD636 pKa = 3.4YY637 pKa = 9.62GQDD640 pKa = 3.16VNNLEE645 pKa = 4.33EE646 pKa = 4.36SVSVGGDD653 pKa = 3.09QTSYY657 pKa = 11.61SFDD660 pKa = 3.46NLASGTWYY668 pKa = 10.56FKK670 pKa = 10.88IEE672 pKa = 4.14VVDD675 pKa = 3.95TNGLSSPPSNPVSTQVQQ692 pKa = 3.28
Molecular weight: 74.73 kDa
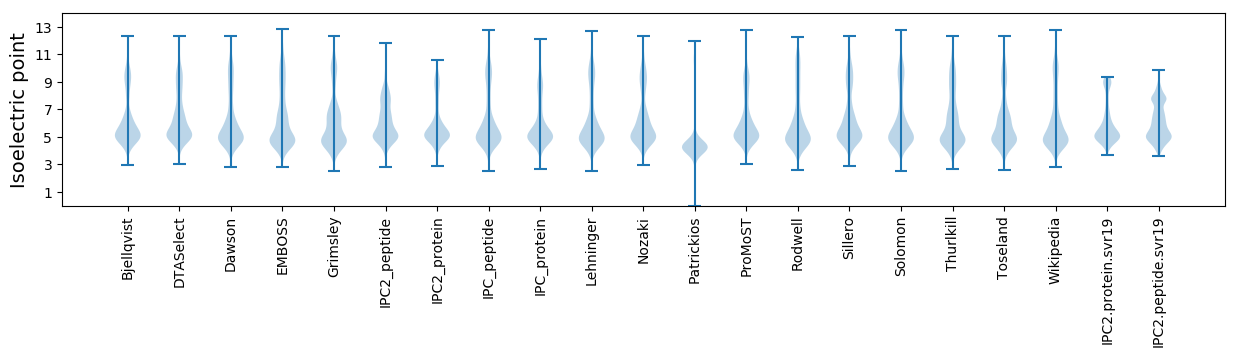
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5YIL3|W5YIL3_9ALTE 3-hydroxyacyl-CoA dehydrogenase OS=Marinobacter similis OX=1420916 GN=AU14_10575 PE=3 SV=1
MM1 pKa = 7.91DD2 pKa = 3.74WMKK5 pKa = 10.86EE6 pKa = 3.59LAALPKK12 pKa = 9.44TLRR15 pKa = 11.84ATLIAALVLLVALILVNQPLQTGSAPQGIVSFQMAGTADD54 pKa = 3.39QAHH57 pKa = 7.27AILRR61 pKa = 11.84SWRR64 pKa = 11.84SGGEE68 pKa = 3.72LWAHH72 pKa = 6.18ISLWLDD78 pKa = 3.11FLFIPAYY85 pKa = 9.89VLALIQLTRR94 pKa = 11.84HH95 pKa = 5.58FTRR98 pKa = 11.84DD99 pKa = 3.06RR100 pKa = 11.84PGVRR104 pKa = 11.84EE105 pKa = 3.84RR106 pKa = 11.84MVARR110 pKa = 11.84WIRR113 pKa = 11.84ALFVAAGLSDD123 pKa = 3.01ITEE126 pKa = 4.65NILLLNNFNPPTDD139 pKa = 3.8VVSLSATICALIKK152 pKa = 10.09YY153 pKa = 7.44TGLTLGIAGLVIVRR167 pKa = 11.84ASRR170 pKa = 11.84RR171 pKa = 11.84HH172 pKa = 5.36PLAHH176 pKa = 6.6GG177 pKa = 3.74
MM1 pKa = 7.91DD2 pKa = 3.74WMKK5 pKa = 10.86EE6 pKa = 3.59LAALPKK12 pKa = 9.44TLRR15 pKa = 11.84ATLIAALVLLVALILVNQPLQTGSAPQGIVSFQMAGTADD54 pKa = 3.39QAHH57 pKa = 7.27AILRR61 pKa = 11.84SWRR64 pKa = 11.84SGGEE68 pKa = 3.72LWAHH72 pKa = 6.18ISLWLDD78 pKa = 3.11FLFIPAYY85 pKa = 9.89VLALIQLTRR94 pKa = 11.84HH95 pKa = 5.58FTRR98 pKa = 11.84DD99 pKa = 3.06RR100 pKa = 11.84PGVRR104 pKa = 11.84EE105 pKa = 3.84RR106 pKa = 11.84MVARR110 pKa = 11.84WIRR113 pKa = 11.84ALFVAAGLSDD123 pKa = 3.01ITEE126 pKa = 4.65NILLLNNFNPPTDD139 pKa = 3.8VVSLSATICALIKK152 pKa = 10.09YY153 pKa = 7.44TGLTLGIAGLVIVRR167 pKa = 11.84ASRR170 pKa = 11.84RR171 pKa = 11.84HH172 pKa = 5.36PLAHH176 pKa = 6.6GG177 pKa = 3.74
Molecular weight: 19.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
735979 |
30 |
1651 |
273.4 |
30.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.861 ± 0.053 | 0.927 ± 0.017 |
5.841 ± 0.041 | 6.547 ± 0.049 |
3.819 ± 0.037 | 7.738 ± 0.042 |
2.137 ± 0.026 | 5.158 ± 0.042 |
3.674 ± 0.043 | 10.626 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.583 ± 0.024 | 3.313 ± 0.03 |
4.617 ± 0.029 | 4.147 ± 0.038 |
6.319 ± 0.047 | 6.198 ± 0.038 |
5.242 ± 0.042 | 7.391 ± 0.035 |
1.347 ± 0.022 | 2.514 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
