
archaeon D22
Taxonomy: cellular organisms; Archaea; unclassified Archaea
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
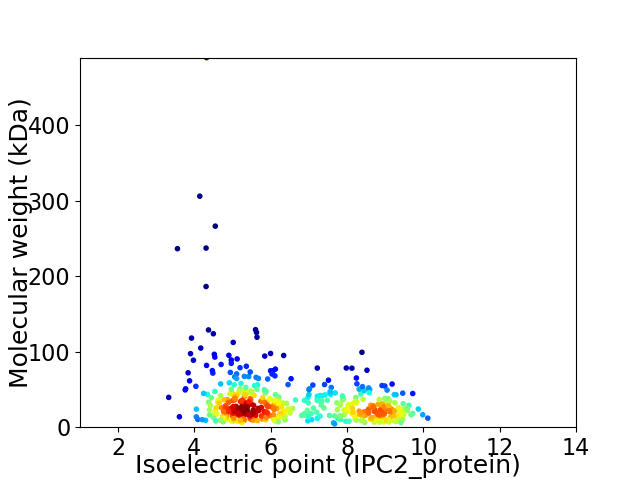
Virtual 2D-PAGE plot for 474 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A202DFH1|A0A202DFH1_9ARCH Uncharacterized protein OS=archaeon D22 OX=1932700 GN=BVX95_01350 PE=4 SV=1
MM1 pKa = 7.34KK2 pKa = 10.34KK3 pKa = 9.74IGIMMLIFATLMIAGCSSPEE23 pKa = 3.68ASVCGDD29 pKa = 3.9GVCSFLEE36 pKa = 4.04KK37 pKa = 10.97SKK39 pKa = 11.18DD40 pKa = 3.65NCPMDD45 pKa = 4.44CKK47 pKa = 10.9ARR49 pKa = 11.84APDD52 pKa = 3.63LSLDD56 pKa = 3.53VEE58 pKa = 4.56FDD60 pKa = 3.17GGTVDD65 pKa = 4.7AYY67 pKa = 11.51SDD69 pKa = 3.98LVLDD73 pKa = 3.33YY74 pKa = 10.18TIRR77 pKa = 11.84NRR79 pKa = 11.84GDD81 pKa = 3.27LATKK85 pKa = 9.72FVVEE89 pKa = 4.4FDD91 pKa = 4.16VEE93 pKa = 4.18RR94 pKa = 11.84LDD96 pKa = 3.98EE97 pKa = 4.39NEE99 pKa = 3.98EE100 pKa = 3.85YY101 pKa = 10.65SYY103 pKa = 11.82GRR105 pKa = 11.84GYY107 pKa = 10.71SRR109 pKa = 11.84QEE111 pKa = 3.41PVMLGEE117 pKa = 4.91GEE119 pKa = 4.21NFNDD123 pKa = 5.21KK124 pKa = 11.21ANFWIRR130 pKa = 11.84EE131 pKa = 3.95EE132 pKa = 4.16GNYY135 pKa = 9.62RR136 pKa = 11.84ISMKK140 pKa = 9.83TYY142 pKa = 9.06PVNEE146 pKa = 3.49RR147 pKa = 11.84DD148 pKa = 4.8SFNEE152 pKa = 3.83DD153 pKa = 2.61NSYY156 pKa = 10.73EE157 pKa = 4.46AIVSAAKK164 pKa = 9.91PSYY167 pKa = 9.37PEE169 pKa = 3.94YY170 pKa = 10.31IIEE173 pKa = 4.11EE174 pKa = 4.36NISAFEE180 pKa = 3.84YY181 pKa = 10.51QYY183 pKa = 10.34TRR185 pKa = 11.84KK186 pKa = 8.68WDD188 pKa = 3.39NGIDD192 pKa = 3.71FDD194 pKa = 4.96EE195 pKa = 6.11IEE197 pKa = 5.18AEE199 pKa = 4.15STYY202 pKa = 10.5NAQYY206 pKa = 10.68EE207 pKa = 4.44YY208 pKa = 11.25VDD210 pKa = 3.99EE211 pKa = 4.74YY212 pKa = 11.36EE213 pKa = 4.57NSAQIDD219 pKa = 4.35LEE221 pKa = 4.41VTTYY225 pKa = 10.71GQDD228 pKa = 2.83ITVEE232 pKa = 3.79ARR234 pKa = 11.84DD235 pKa = 3.76FFMSYY240 pKa = 10.78SSFEE244 pKa = 4.0AEE246 pKa = 4.35NYY248 pKa = 9.92KK249 pKa = 10.9GDD251 pKa = 3.76IIFIDD256 pKa = 4.26GEE258 pKa = 4.74DD259 pKa = 3.69YY260 pKa = 11.63GDD262 pKa = 3.32MQFFEE267 pKa = 5.13VVWPSNDD274 pKa = 2.84KK275 pKa = 10.63LVRR278 pKa = 11.84LFIHH282 pKa = 6.58FNEE285 pKa = 4.55KK286 pKa = 10.47EE287 pKa = 3.77MDD289 pKa = 3.28GDD291 pKa = 4.09YY292 pKa = 10.94IEE294 pKa = 5.39EE295 pKa = 4.12FVDD298 pKa = 3.98KK299 pKa = 10.85YY300 pKa = 10.76LAKK303 pKa = 10.2FDD305 pKa = 4.22SSVEE309 pKa = 3.89YY310 pKa = 11.44NMLCGDD316 pKa = 4.84NKK318 pKa = 10.49CQYY321 pKa = 11.51NNIFLQQSEE330 pKa = 5.14TYY332 pKa = 9.0STEE335 pKa = 4.07FNDD338 pKa = 3.84NNVHH342 pKa = 6.57EE343 pKa = 4.6ISVLNISDD351 pKa = 3.46TGAQITIDD359 pKa = 3.73DD360 pKa = 3.69DD361 pKa = 4.47TYY363 pKa = 11.7FEE365 pKa = 4.15FSEE368 pKa = 3.97IGEE371 pKa = 4.29VVDD374 pKa = 4.78YY375 pKa = 10.89NGQSIEE381 pKa = 4.18LRR383 pKa = 11.84NLAPTEE389 pKa = 3.77IEE391 pKa = 4.86FIDD394 pKa = 4.03FTSSNVEE401 pKa = 3.74IVVATINEE409 pKa = 4.02EE410 pKa = 4.29GNLKK414 pKa = 10.32NYY416 pKa = 10.41EE417 pKa = 4.21EE418 pKa = 5.35IEE420 pKa = 4.23VQLGKK425 pKa = 10.44EE426 pKa = 3.81YY427 pKa = 10.95SVNFDD432 pKa = 3.4EE433 pKa = 5.49GNYY436 pKa = 9.95SIKK439 pKa = 10.51LIKK442 pKa = 10.38FKK444 pKa = 11.07EE445 pKa = 4.14DD446 pKa = 3.17VAVIEE451 pKa = 4.37VNDD454 pKa = 4.08DD455 pKa = 3.46KK456 pKa = 11.65EE457 pKa = 5.15SIILHH462 pKa = 5.39EE463 pKa = 4.28EE464 pKa = 4.05EE465 pKa = 5.26IIGGLYY471 pKa = 10.31VRR473 pKa = 11.84VDD475 pKa = 4.67DD476 pKa = 5.32IDD478 pKa = 4.16IDD480 pKa = 4.02FDD482 pKa = 3.99TEE484 pKa = 4.06TRR486 pKa = 11.84PEE488 pKa = 3.83AVILNIGEE496 pKa = 4.3NYY498 pKa = 8.64QACPSDD504 pKa = 4.26CAPMVAQEE512 pKa = 4.3EE513 pKa = 4.41PMPVSSSGGSAGEE526 pKa = 4.22SGPVMGSEE534 pKa = 4.43VEE536 pKa = 3.98LSEE539 pKa = 4.49
MM1 pKa = 7.34KK2 pKa = 10.34KK3 pKa = 9.74IGIMMLIFATLMIAGCSSPEE23 pKa = 3.68ASVCGDD29 pKa = 3.9GVCSFLEE36 pKa = 4.04KK37 pKa = 10.97SKK39 pKa = 11.18DD40 pKa = 3.65NCPMDD45 pKa = 4.44CKK47 pKa = 10.9ARR49 pKa = 11.84APDD52 pKa = 3.63LSLDD56 pKa = 3.53VEE58 pKa = 4.56FDD60 pKa = 3.17GGTVDD65 pKa = 4.7AYY67 pKa = 11.51SDD69 pKa = 3.98LVLDD73 pKa = 3.33YY74 pKa = 10.18TIRR77 pKa = 11.84NRR79 pKa = 11.84GDD81 pKa = 3.27LATKK85 pKa = 9.72FVVEE89 pKa = 4.4FDD91 pKa = 4.16VEE93 pKa = 4.18RR94 pKa = 11.84LDD96 pKa = 3.98EE97 pKa = 4.39NEE99 pKa = 3.98EE100 pKa = 3.85YY101 pKa = 10.65SYY103 pKa = 11.82GRR105 pKa = 11.84GYY107 pKa = 10.71SRR109 pKa = 11.84QEE111 pKa = 3.41PVMLGEE117 pKa = 4.91GEE119 pKa = 4.21NFNDD123 pKa = 5.21KK124 pKa = 11.21ANFWIRR130 pKa = 11.84EE131 pKa = 3.95EE132 pKa = 4.16GNYY135 pKa = 9.62RR136 pKa = 11.84ISMKK140 pKa = 9.83TYY142 pKa = 9.06PVNEE146 pKa = 3.49RR147 pKa = 11.84DD148 pKa = 4.8SFNEE152 pKa = 3.83DD153 pKa = 2.61NSYY156 pKa = 10.73EE157 pKa = 4.46AIVSAAKK164 pKa = 9.91PSYY167 pKa = 9.37PEE169 pKa = 3.94YY170 pKa = 10.31IIEE173 pKa = 4.11EE174 pKa = 4.36NISAFEE180 pKa = 3.84YY181 pKa = 10.51QYY183 pKa = 10.34TRR185 pKa = 11.84KK186 pKa = 8.68WDD188 pKa = 3.39NGIDD192 pKa = 3.71FDD194 pKa = 4.96EE195 pKa = 6.11IEE197 pKa = 5.18AEE199 pKa = 4.15STYY202 pKa = 10.5NAQYY206 pKa = 10.68EE207 pKa = 4.44YY208 pKa = 11.25VDD210 pKa = 3.99EE211 pKa = 4.74YY212 pKa = 11.36EE213 pKa = 4.57NSAQIDD219 pKa = 4.35LEE221 pKa = 4.41VTTYY225 pKa = 10.71GQDD228 pKa = 2.83ITVEE232 pKa = 3.79ARR234 pKa = 11.84DD235 pKa = 3.76FFMSYY240 pKa = 10.78SSFEE244 pKa = 4.0AEE246 pKa = 4.35NYY248 pKa = 9.92KK249 pKa = 10.9GDD251 pKa = 3.76IIFIDD256 pKa = 4.26GEE258 pKa = 4.74DD259 pKa = 3.69YY260 pKa = 11.63GDD262 pKa = 3.32MQFFEE267 pKa = 5.13VVWPSNDD274 pKa = 2.84KK275 pKa = 10.63LVRR278 pKa = 11.84LFIHH282 pKa = 6.58FNEE285 pKa = 4.55KK286 pKa = 10.47EE287 pKa = 3.77MDD289 pKa = 3.28GDD291 pKa = 4.09YY292 pKa = 10.94IEE294 pKa = 5.39EE295 pKa = 4.12FVDD298 pKa = 3.98KK299 pKa = 10.85YY300 pKa = 10.76LAKK303 pKa = 10.2FDD305 pKa = 4.22SSVEE309 pKa = 3.89YY310 pKa = 11.44NMLCGDD316 pKa = 4.84NKK318 pKa = 10.49CQYY321 pKa = 11.51NNIFLQQSEE330 pKa = 5.14TYY332 pKa = 9.0STEE335 pKa = 4.07FNDD338 pKa = 3.84NNVHH342 pKa = 6.57EE343 pKa = 4.6ISVLNISDD351 pKa = 3.46TGAQITIDD359 pKa = 3.73DD360 pKa = 3.69DD361 pKa = 4.47TYY363 pKa = 11.7FEE365 pKa = 4.15FSEE368 pKa = 3.97IGEE371 pKa = 4.29VVDD374 pKa = 4.78YY375 pKa = 10.89NGQSIEE381 pKa = 4.18LRR383 pKa = 11.84NLAPTEE389 pKa = 3.77IEE391 pKa = 4.86FIDD394 pKa = 4.03FTSSNVEE401 pKa = 3.74IVVATINEE409 pKa = 4.02EE410 pKa = 4.29GNLKK414 pKa = 10.32NYY416 pKa = 10.41EE417 pKa = 4.21EE418 pKa = 5.35IEE420 pKa = 4.23VQLGKK425 pKa = 10.44EE426 pKa = 3.81YY427 pKa = 10.95SVNFDD432 pKa = 3.4EE433 pKa = 5.49GNYY436 pKa = 9.95SIKK439 pKa = 10.51LIKK442 pKa = 10.38FKK444 pKa = 11.07EE445 pKa = 4.14DD446 pKa = 3.17VAVIEE451 pKa = 4.37VNDD454 pKa = 4.08DD455 pKa = 3.46KK456 pKa = 11.65EE457 pKa = 5.15SIILHH462 pKa = 5.39EE463 pKa = 4.28EE464 pKa = 4.05EE465 pKa = 5.26IIGGLYY471 pKa = 10.31VRR473 pKa = 11.84VDD475 pKa = 4.67DD476 pKa = 5.32IDD478 pKa = 4.16IDD480 pKa = 4.02FDD482 pKa = 3.99TEE484 pKa = 4.06TRR486 pKa = 11.84PEE488 pKa = 3.83AVILNIGEE496 pKa = 4.3NYY498 pKa = 8.64QACPSDD504 pKa = 4.26CAPMVAQEE512 pKa = 4.3EE513 pKa = 4.41PMPVSSSGGSAGEE526 pKa = 4.22SGPVMGSEE534 pKa = 4.43VEE536 pKa = 3.98LSEE539 pKa = 4.49
Molecular weight: 61.37 kDa
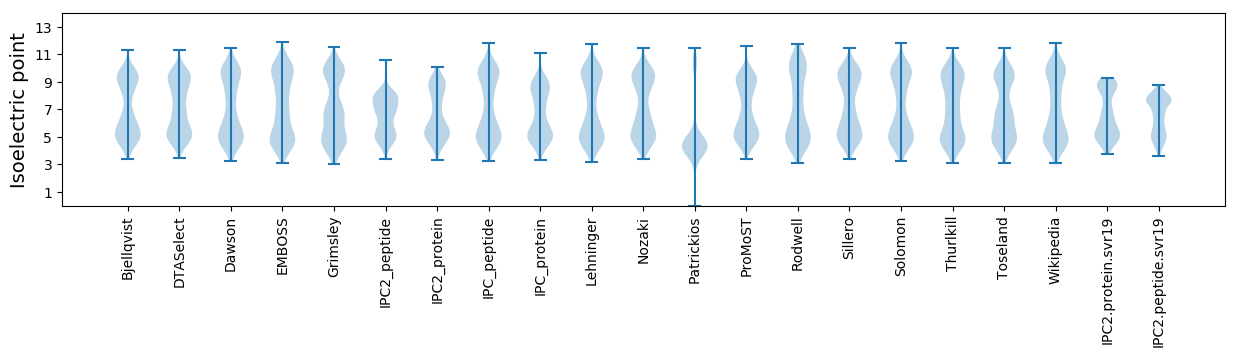
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A202DG21|A0A202DG21_9ARCH Uncharacterized protein OS=archaeon D22 OX=1932700 GN=BVX95_00645 PE=4 SV=1
MM1 pKa = 7.46TFFTSFILGLLTPLTAVCVLPLVPGFLVYY30 pKa = 10.8LSNQIGTQTSKK41 pKa = 10.97LRR43 pKa = 11.84TLMLGVIMTLGVILFMGLIGVVFSLLLQISLTNVIGIISPIAFGTLAIFSILLIFNVDD101 pKa = 2.75IGKK104 pKa = 9.32FLPKK108 pKa = 10.34FKK110 pKa = 11.06SPVAKK115 pKa = 10.5SPILSSFLFGFFFGAIVLPCNPLFIAAMFSRR146 pKa = 11.84ALTFTDD152 pKa = 3.17LTMNIINFIFFGFGMGFPLIFLSAVSAATGTKK184 pKa = 9.84LVSFLSKK191 pKa = 10.59NKK193 pKa = 10.22RR194 pKa = 11.84ILNLISGVIMLGISIYY210 pKa = 11.42YY211 pKa = 7.82MFFVFRR217 pKa = 11.84ILRR220 pKa = 3.85
MM1 pKa = 7.46TFFTSFILGLLTPLTAVCVLPLVPGFLVYY30 pKa = 10.8LSNQIGTQTSKK41 pKa = 10.97LRR43 pKa = 11.84TLMLGVIMTLGVILFMGLIGVVFSLLLQISLTNVIGIISPIAFGTLAIFSILLIFNVDD101 pKa = 2.75IGKK104 pKa = 9.32FLPKK108 pKa = 10.34FKK110 pKa = 11.06SPVAKK115 pKa = 10.5SPILSSFLFGFFFGAIVLPCNPLFIAAMFSRR146 pKa = 11.84ALTFTDD152 pKa = 3.17LTMNIINFIFFGFGMGFPLIFLSAVSAATGTKK184 pKa = 9.84LVSFLSKK191 pKa = 10.59NKK193 pKa = 10.22RR194 pKa = 11.84ILNLISGVIMLGISIYY210 pKa = 11.42YY211 pKa = 7.82MFFVFRR217 pKa = 11.84ILRR220 pKa = 3.85
Molecular weight: 24.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
147978 |
39 |
4423 |
312.2 |
35.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.522 ± 0.124 | 1.008 ± 0.052 |
6.281 ± 0.152 | 7.623 ± 0.172 |
4.885 ± 0.115 | 6.252 ± 0.127 |
1.693 ± 0.05 | 8.492 ± 0.135 |
8.504 ± 0.249 | 8.907 ± 0.156 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.224 ± 0.075 | 5.832 ± 0.21 |
3.108 ± 0.064 | 2.628 ± 0.066 |
3.442 ± 0.113 | 7.073 ± 0.177 |
5.24 ± 0.178 | 6.546 ± 0.098 |
0.762 ± 0.038 | 3.978 ± 0.102 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
