
Capybara microvirus Cap1_SP_90
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.72
Get precalculated fractions of proteins
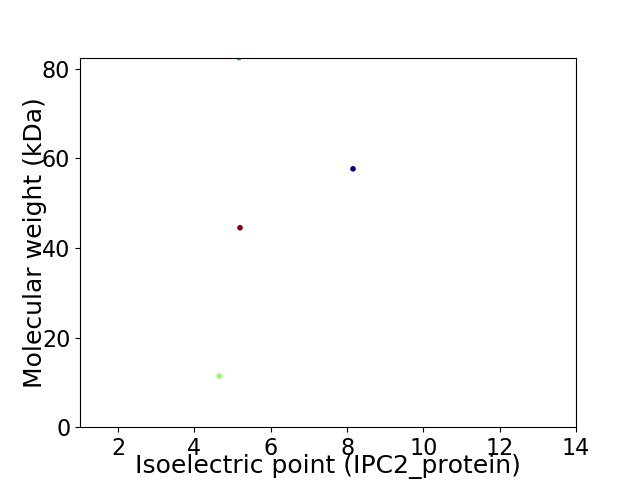
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W418|A0A4P8W418_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_90 OX=2584797 PE=4 SV=1
MM1 pKa = 7.37FPTVEE6 pKa = 4.44KK7 pKa = 9.41FQCSRR12 pKa = 11.84GCPVYY17 pKa = 10.05ATVDD21 pKa = 3.62RR22 pKa = 11.84EE23 pKa = 4.18VLIDD27 pKa = 3.74SDD29 pKa = 4.1GLAVPIIEE37 pKa = 4.41KK38 pKa = 9.52TVIDD42 pKa = 3.93MTTEE46 pKa = 3.82EE47 pKa = 4.69SIKK50 pKa = 10.99SLGTGLSLKK59 pKa = 10.43DD60 pKa = 3.45ALEE63 pKa = 4.32AGISIDD69 pKa = 3.75RR70 pKa = 11.84VNTQVLSTSRR80 pKa = 11.84ADD82 pKa = 3.55NLEE85 pKa = 4.16KK86 pKa = 10.78YY87 pKa = 10.41VEE89 pKa = 5.02LINKK93 pKa = 9.81DD94 pKa = 2.92IEE96 pKa = 4.31RR97 pKa = 11.84TLNEE101 pKa = 4.11SKK103 pKa = 11.05
MM1 pKa = 7.37FPTVEE6 pKa = 4.44KK7 pKa = 9.41FQCSRR12 pKa = 11.84GCPVYY17 pKa = 10.05ATVDD21 pKa = 3.62RR22 pKa = 11.84EE23 pKa = 4.18VLIDD27 pKa = 3.74SDD29 pKa = 4.1GLAVPIIEE37 pKa = 4.41KK38 pKa = 9.52TVIDD42 pKa = 3.93MTTEE46 pKa = 3.82EE47 pKa = 4.69SIKK50 pKa = 10.99SLGTGLSLKK59 pKa = 10.43DD60 pKa = 3.45ALEE63 pKa = 4.32AGISIDD69 pKa = 3.75RR70 pKa = 11.84VNTQVLSTSRR80 pKa = 11.84ADD82 pKa = 3.55NLEE85 pKa = 4.16KK86 pKa = 10.78YY87 pKa = 10.41VEE89 pKa = 5.02LINKK93 pKa = 9.81DD94 pKa = 2.92IEE96 pKa = 4.31RR97 pKa = 11.84TLNEE101 pKa = 4.11SKK103 pKa = 11.05
Molecular weight: 11.38 kDa
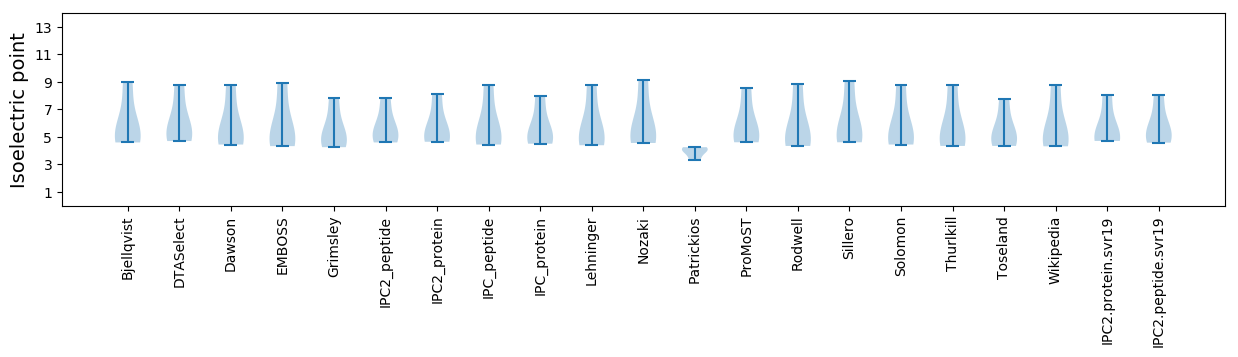
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVN4|A0A4V1FVN4_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_90 OX=2584797 PE=4 SV=1
MM1 pKa = 7.97CYY3 pKa = 9.99NPRR6 pKa = 11.84VIRR9 pKa = 11.84NPTKK13 pKa = 10.83DD14 pKa = 3.98LVPTDD19 pKa = 3.68ALQITVSCGEE29 pKa = 4.08CEE31 pKa = 3.74EE32 pKa = 4.6CRR34 pKa = 11.84NHH36 pKa = 6.65RR37 pKa = 11.84RR38 pKa = 11.84NEE40 pKa = 4.08MVFRR44 pKa = 11.84TYY46 pKa = 10.9EE47 pKa = 4.25QMLEE51 pKa = 4.07DD52 pKa = 3.64QKK54 pKa = 11.4KK55 pKa = 10.83GYY57 pKa = 8.55IQCFFTLTYY66 pKa = 11.07NDD68 pKa = 3.82DD69 pKa = 4.0CLPHH73 pKa = 6.35FKK75 pKa = 10.34CSSYY79 pKa = 10.95NIPCFSPDD87 pKa = 4.06DD88 pKa = 3.47ISAFRR93 pKa = 11.84GHH95 pKa = 6.43LQDD98 pKa = 4.29YY99 pKa = 10.69LLDD102 pKa = 4.23RR103 pKa = 11.84YY104 pKa = 10.07GIKK107 pKa = 10.38SSTYY111 pKa = 9.97LICAEE116 pKa = 4.38YY117 pKa = 10.75GSNTQRR123 pKa = 11.84PHH125 pKa = 4.14YY126 pKa = 9.23HH127 pKa = 7.15CYY129 pKa = 9.2FACPSKK135 pKa = 11.1GVITPRR141 pKa = 11.84FTKK144 pKa = 9.29KK145 pKa = 9.17TGITSEE151 pKa = 4.36SNLPVLTGSILHH163 pKa = 6.49SIVKK167 pKa = 9.27EE168 pKa = 3.66CWTYY172 pKa = 11.25GFLFPRR178 pKa = 11.84FPQGGRR184 pKa = 11.84DD185 pKa = 3.39STGYY189 pKa = 7.94NHH191 pKa = 7.33KK192 pKa = 10.46PFVLTGDD199 pKa = 3.33VTGAAKK205 pKa = 10.61YY206 pKa = 8.54IAKK209 pKa = 9.49YY210 pKa = 8.82VCKK213 pKa = 10.44DD214 pKa = 3.17LALLEE219 pKa = 4.66RR220 pKa = 11.84PDD222 pKa = 3.56VQQFIAEE229 pKa = 4.87FKK231 pKa = 10.44NKK233 pKa = 10.27LNGKK237 pKa = 8.82HH238 pKa = 6.92DD239 pKa = 3.65IYY241 pKa = 11.51KK242 pKa = 10.34KK243 pKa = 8.48FQKK246 pKa = 10.19VSSLMRR252 pKa = 11.84CSQGFGKK259 pKa = 9.91CICDD263 pKa = 3.38KK264 pKa = 10.94VKK266 pKa = 10.98SVDD269 pKa = 3.93DD270 pKa = 3.79IKK272 pKa = 11.61NGVTTNLYY280 pKa = 5.9PTHH283 pKa = 7.05KK284 pKa = 9.96IAVPQYY290 pKa = 9.64IVRR293 pKa = 11.84KK294 pKa = 6.81LTRR297 pKa = 11.84VAHH300 pKa = 5.68YY301 pKa = 9.97IKK303 pKa = 10.55EE304 pKa = 4.16DD305 pKa = 3.32ASGFKK310 pKa = 10.1YY311 pKa = 10.54LKK313 pKa = 10.06PKK315 pKa = 9.84VRR317 pKa = 11.84YY318 pKa = 8.9EE319 pKa = 3.59LTEE322 pKa = 4.12FGKK325 pKa = 10.43EE326 pKa = 4.03VYY328 pKa = 10.01KK329 pKa = 10.82EE330 pKa = 4.24SIKK333 pKa = 10.81HH334 pKa = 5.13QIKK337 pKa = 10.61NLTKK341 pKa = 10.41SLHH344 pKa = 6.5DD345 pKa = 4.04WNPSIDD351 pKa = 3.71AQSIATYY358 pKa = 10.42AVVYY362 pKa = 9.34RR363 pKa = 11.84NRR365 pKa = 11.84MNPRR369 pKa = 11.84HH370 pKa = 5.5STIKK374 pKa = 10.74DD375 pKa = 3.09AFTCEE380 pKa = 3.81LTSADD385 pKa = 4.01FFTGLEE391 pKa = 4.11SLSRR395 pKa = 11.84MMSKK399 pKa = 10.66ALPYY403 pKa = 10.45KK404 pKa = 10.03FAHH407 pKa = 6.65LDD409 pKa = 3.72YY410 pKa = 11.2VKK412 pKa = 10.61LPQSPLVNDD421 pKa = 3.67DD422 pKa = 3.57VYY424 pKa = 11.45YY425 pKa = 10.93HH426 pKa = 7.34LDD428 pKa = 2.94LGYY431 pKa = 10.64RR432 pKa = 11.84FNDD435 pKa = 4.39FKK437 pKa = 11.23CFEE440 pKa = 4.58GFDD443 pKa = 4.89DD444 pKa = 5.22IYY446 pKa = 11.45DD447 pKa = 3.81SYY449 pKa = 12.2VEE451 pKa = 4.04YY452 pKa = 11.11NNKK455 pKa = 9.3KK456 pKa = 10.11KK457 pKa = 10.76KK458 pKa = 10.39EE459 pKa = 3.9NLEE462 pKa = 3.91LKK464 pKa = 9.95AAHH467 pKa = 5.98AQYY470 pKa = 10.55IKK472 pKa = 10.57SLKK475 pKa = 9.05EE476 pKa = 4.04HH477 pKa = 6.96FSHH480 pKa = 7.42TEE482 pKa = 3.49NNNNNKK488 pKa = 9.05YY489 pKa = 10.27RR490 pKa = 11.84SLCFRR495 pKa = 11.84QQ496 pKa = 3.3
MM1 pKa = 7.97CYY3 pKa = 9.99NPRR6 pKa = 11.84VIRR9 pKa = 11.84NPTKK13 pKa = 10.83DD14 pKa = 3.98LVPTDD19 pKa = 3.68ALQITVSCGEE29 pKa = 4.08CEE31 pKa = 3.74EE32 pKa = 4.6CRR34 pKa = 11.84NHH36 pKa = 6.65RR37 pKa = 11.84RR38 pKa = 11.84NEE40 pKa = 4.08MVFRR44 pKa = 11.84TYY46 pKa = 10.9EE47 pKa = 4.25QMLEE51 pKa = 4.07DD52 pKa = 3.64QKK54 pKa = 11.4KK55 pKa = 10.83GYY57 pKa = 8.55IQCFFTLTYY66 pKa = 11.07NDD68 pKa = 3.82DD69 pKa = 4.0CLPHH73 pKa = 6.35FKK75 pKa = 10.34CSSYY79 pKa = 10.95NIPCFSPDD87 pKa = 4.06DD88 pKa = 3.47ISAFRR93 pKa = 11.84GHH95 pKa = 6.43LQDD98 pKa = 4.29YY99 pKa = 10.69LLDD102 pKa = 4.23RR103 pKa = 11.84YY104 pKa = 10.07GIKK107 pKa = 10.38SSTYY111 pKa = 9.97LICAEE116 pKa = 4.38YY117 pKa = 10.75GSNTQRR123 pKa = 11.84PHH125 pKa = 4.14YY126 pKa = 9.23HH127 pKa = 7.15CYY129 pKa = 9.2FACPSKK135 pKa = 11.1GVITPRR141 pKa = 11.84FTKK144 pKa = 9.29KK145 pKa = 9.17TGITSEE151 pKa = 4.36SNLPVLTGSILHH163 pKa = 6.49SIVKK167 pKa = 9.27EE168 pKa = 3.66CWTYY172 pKa = 11.25GFLFPRR178 pKa = 11.84FPQGGRR184 pKa = 11.84DD185 pKa = 3.39STGYY189 pKa = 7.94NHH191 pKa = 7.33KK192 pKa = 10.46PFVLTGDD199 pKa = 3.33VTGAAKK205 pKa = 10.61YY206 pKa = 8.54IAKK209 pKa = 9.49YY210 pKa = 8.82VCKK213 pKa = 10.44DD214 pKa = 3.17LALLEE219 pKa = 4.66RR220 pKa = 11.84PDD222 pKa = 3.56VQQFIAEE229 pKa = 4.87FKK231 pKa = 10.44NKK233 pKa = 10.27LNGKK237 pKa = 8.82HH238 pKa = 6.92DD239 pKa = 3.65IYY241 pKa = 11.51KK242 pKa = 10.34KK243 pKa = 8.48FQKK246 pKa = 10.19VSSLMRR252 pKa = 11.84CSQGFGKK259 pKa = 9.91CICDD263 pKa = 3.38KK264 pKa = 10.94VKK266 pKa = 10.98SVDD269 pKa = 3.93DD270 pKa = 3.79IKK272 pKa = 11.61NGVTTNLYY280 pKa = 5.9PTHH283 pKa = 7.05KK284 pKa = 9.96IAVPQYY290 pKa = 9.64IVRR293 pKa = 11.84KK294 pKa = 6.81LTRR297 pKa = 11.84VAHH300 pKa = 5.68YY301 pKa = 9.97IKK303 pKa = 10.55EE304 pKa = 4.16DD305 pKa = 3.32ASGFKK310 pKa = 10.1YY311 pKa = 10.54LKK313 pKa = 10.06PKK315 pKa = 9.84VRR317 pKa = 11.84YY318 pKa = 8.9EE319 pKa = 3.59LTEE322 pKa = 4.12FGKK325 pKa = 10.43EE326 pKa = 4.03VYY328 pKa = 10.01KK329 pKa = 10.82EE330 pKa = 4.24SIKK333 pKa = 10.81HH334 pKa = 5.13QIKK337 pKa = 10.61NLTKK341 pKa = 10.41SLHH344 pKa = 6.5DD345 pKa = 4.04WNPSIDD351 pKa = 3.71AQSIATYY358 pKa = 10.42AVVYY362 pKa = 9.34RR363 pKa = 11.84NRR365 pKa = 11.84MNPRR369 pKa = 11.84HH370 pKa = 5.5STIKK374 pKa = 10.74DD375 pKa = 3.09AFTCEE380 pKa = 3.81LTSADD385 pKa = 4.01FFTGLEE391 pKa = 4.11SLSRR395 pKa = 11.84MMSKK399 pKa = 10.66ALPYY403 pKa = 10.45KK404 pKa = 10.03FAHH407 pKa = 6.65LDD409 pKa = 3.72YY410 pKa = 11.2VKK412 pKa = 10.61LPQSPLVNDD421 pKa = 3.67DD422 pKa = 3.57VYY424 pKa = 11.45YY425 pKa = 10.93HH426 pKa = 7.34LDD428 pKa = 2.94LGYY431 pKa = 10.64RR432 pKa = 11.84FNDD435 pKa = 4.39FKK437 pKa = 11.23CFEE440 pKa = 4.58GFDD443 pKa = 4.89DD444 pKa = 5.22IYY446 pKa = 11.45DD447 pKa = 3.81SYY449 pKa = 12.2VEE451 pKa = 4.04YY452 pKa = 11.11NNKK455 pKa = 9.3KK456 pKa = 10.11KK457 pKa = 10.76KK458 pKa = 10.39EE459 pKa = 3.9NLEE462 pKa = 3.91LKK464 pKa = 9.95AAHH467 pKa = 5.98AQYY470 pKa = 10.55IKK472 pKa = 10.57SLKK475 pKa = 9.05EE476 pKa = 4.04HH477 pKa = 6.96FSHH480 pKa = 7.42TEE482 pKa = 3.49NNNNNKK488 pKa = 9.05YY489 pKa = 10.27RR490 pKa = 11.84SLCFRR495 pKa = 11.84QQ496 pKa = 3.3
Molecular weight: 57.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1739 |
103 |
733 |
434.8 |
48.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.383 ± 1.585 | 1.725 ± 0.771 |
6.21 ± 0.238 | 5.003 ± 0.496 |
4.485 ± 0.538 | 5.808 ± 0.707 |
1.783 ± 0.674 | 5.463 ± 0.579 |
6.153 ± 1.334 | 8.338 ± 0.279 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.358 ± 0.384 | 6.038 ± 0.557 |
4.485 ± 0.542 | 3.968 ± 0.584 |
4.428 ± 0.169 | 8.626 ± 0.718 |
6.671 ± 0.5 | 5.693 ± 0.319 |
0.805 ± 0.238 | 5.578 ± 0.624 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
