
Potato virus H
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Quinvirinae; Carlavirus
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
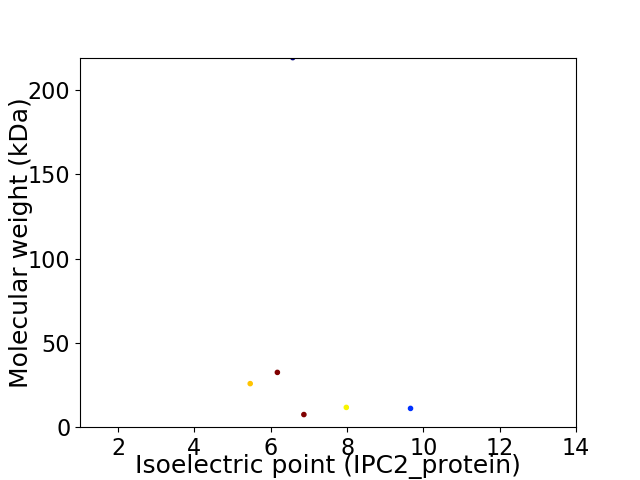
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I6Z8L3|I6Z8L3_9VIRU 26K protein OS=Potato virus H OX=1046402 GN=TGB1 PE=4 SV=1
MM1 pKa = 7.86DD2 pKa = 4.2VLVDD6 pKa = 4.43LLNKK10 pKa = 10.25YY11 pKa = 10.52SFTRR15 pKa = 11.84IHH17 pKa = 6.8SKK19 pKa = 10.29LSSPIVVHH27 pKa = 6.1CVPGAGKK34 pKa = 8.68TSLIRR39 pKa = 11.84EE40 pKa = 4.62LIKK43 pKa = 10.57LDD45 pKa = 3.65SRR47 pKa = 11.84FVAYY51 pKa = 8.36TAGVEE56 pKa = 4.26DD57 pKa = 4.79EE58 pKa = 4.39PHH60 pKa = 7.11LSGRR64 pKa = 11.84WIRR67 pKa = 11.84KK68 pKa = 8.62FEE70 pKa = 4.06GVVDD74 pKa = 3.76EE75 pKa = 4.85GKK77 pKa = 10.54FVILDD82 pKa = 3.89EE83 pKa = 4.35YY84 pKa = 10.8TLLEE88 pKa = 4.34SLPDD92 pKa = 3.33NLFAVFGDD100 pKa = 4.57PIQSNTKK107 pKa = 7.79VVRR110 pKa = 11.84SADD113 pKa = 3.64YY114 pKa = 9.14TCNRR118 pKa = 11.84SKK120 pKa = 11.11RR121 pKa = 11.84FGRR124 pKa = 11.84STALFLRR131 pKa = 11.84EE132 pKa = 4.02LGFDD136 pKa = 3.72VVAEE140 pKa = 4.4ADD142 pKa = 4.03DD143 pKa = 4.07EE144 pKa = 4.75VAVANIYY151 pKa = 10.25QVDD154 pKa = 3.73PVEE157 pKa = 3.87QVVYY161 pKa = 9.74FEE163 pKa = 4.65QEE165 pKa = 4.0VGCLLRR171 pKa = 11.84AHH173 pKa = 6.57HH174 pKa = 6.42VEE176 pKa = 4.19CKK178 pKa = 10.13HH179 pKa = 4.34YY180 pKa = 9.97TEE182 pKa = 5.33IVGQTFEE189 pKa = 4.36KK190 pKa = 9.56VTFVSSEE197 pKa = 4.3SNLSSNRR204 pKa = 11.84VAAYY208 pKa = 10.0QCMTRR213 pKa = 11.84HH214 pKa = 6.02RR215 pKa = 11.84SKK217 pKa = 11.22LLILTPDD224 pKa = 3.21ATFTAAA230 pKa = 4.8
MM1 pKa = 7.86DD2 pKa = 4.2VLVDD6 pKa = 4.43LLNKK10 pKa = 10.25YY11 pKa = 10.52SFTRR15 pKa = 11.84IHH17 pKa = 6.8SKK19 pKa = 10.29LSSPIVVHH27 pKa = 6.1CVPGAGKK34 pKa = 8.68TSLIRR39 pKa = 11.84EE40 pKa = 4.62LIKK43 pKa = 10.57LDD45 pKa = 3.65SRR47 pKa = 11.84FVAYY51 pKa = 8.36TAGVEE56 pKa = 4.26DD57 pKa = 4.79EE58 pKa = 4.39PHH60 pKa = 7.11LSGRR64 pKa = 11.84WIRR67 pKa = 11.84KK68 pKa = 8.62FEE70 pKa = 4.06GVVDD74 pKa = 3.76EE75 pKa = 4.85GKK77 pKa = 10.54FVILDD82 pKa = 3.89EE83 pKa = 4.35YY84 pKa = 10.8TLLEE88 pKa = 4.34SLPDD92 pKa = 3.33NLFAVFGDD100 pKa = 4.57PIQSNTKK107 pKa = 7.79VVRR110 pKa = 11.84SADD113 pKa = 3.64YY114 pKa = 9.14TCNRR118 pKa = 11.84SKK120 pKa = 11.11RR121 pKa = 11.84FGRR124 pKa = 11.84STALFLRR131 pKa = 11.84EE132 pKa = 4.02LGFDD136 pKa = 3.72VVAEE140 pKa = 4.4ADD142 pKa = 4.03DD143 pKa = 4.07EE144 pKa = 4.75VAVANIYY151 pKa = 10.25QVDD154 pKa = 3.73PVEE157 pKa = 3.87QVVYY161 pKa = 9.74FEE163 pKa = 4.65QEE165 pKa = 4.0VGCLLRR171 pKa = 11.84AHH173 pKa = 6.57HH174 pKa = 6.42VEE176 pKa = 4.19CKK178 pKa = 10.13HH179 pKa = 4.34YY180 pKa = 9.97TEE182 pKa = 5.33IVGQTFEE189 pKa = 4.36KK190 pKa = 9.56VTFVSSEE197 pKa = 4.3SNLSSNRR204 pKa = 11.84VAAYY208 pKa = 10.0QCMTRR213 pKa = 11.84HH214 pKa = 6.02RR215 pKa = 11.84SKK217 pKa = 11.22LLILTPDD224 pKa = 3.21ATFTAAA230 pKa = 4.8
Molecular weight: 25.86 kDa
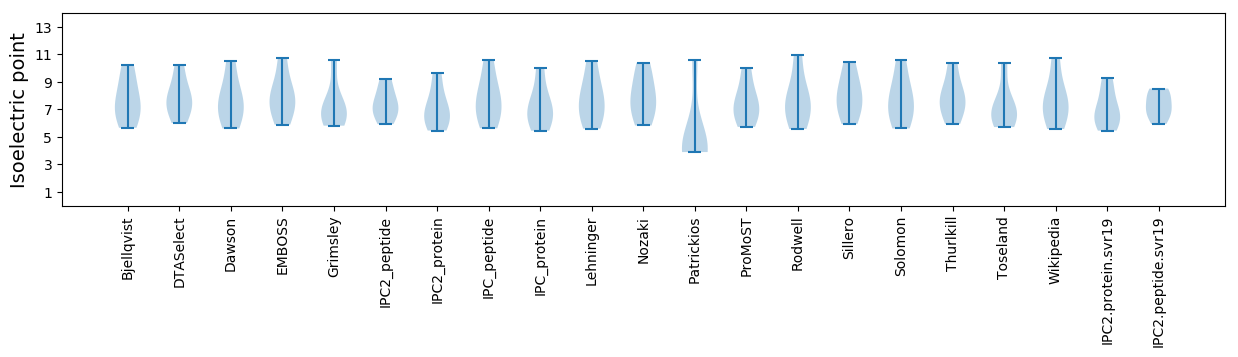
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I6YW94|I6YW94_9VIRU Capsid protein OS=Potato virus H OX=1046402 GN=CP PE=3 SV=1
MM1 pKa = 7.61RR2 pKa = 11.84LSSKK6 pKa = 8.49VLRR9 pKa = 11.84KK10 pKa = 10.16VLVGVFKK17 pKa = 10.57RR18 pKa = 11.84HH19 pKa = 5.67YY20 pKa = 9.22GLGPRR25 pKa = 11.84RR26 pKa = 11.84EE27 pKa = 4.12YY28 pKa = 11.5DD29 pKa = 4.04DD30 pKa = 3.59IASVIISKK38 pKa = 10.64LEE40 pKa = 3.87PPEE43 pKa = 4.52VGLSTYY49 pKa = 10.74AKK51 pKa = 9.8RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84AKK56 pKa = 10.57KK57 pKa = 9.97LGRR60 pKa = 11.84CPRR63 pKa = 11.84CYY65 pKa = 9.88RR66 pKa = 11.84VIGLSQKK73 pKa = 10.51CDD75 pKa = 3.09GFKK78 pKa = 10.42CVPGISYY85 pKa = 9.83NRR87 pKa = 11.84KK88 pKa = 7.83VEE90 pKa = 3.86QNIRR94 pKa = 11.84YY95 pKa = 9.73GDD97 pKa = 3.32
MM1 pKa = 7.61RR2 pKa = 11.84LSSKK6 pKa = 8.49VLRR9 pKa = 11.84KK10 pKa = 10.16VLVGVFKK17 pKa = 10.57RR18 pKa = 11.84HH19 pKa = 5.67YY20 pKa = 9.22GLGPRR25 pKa = 11.84RR26 pKa = 11.84EE27 pKa = 4.12YY28 pKa = 11.5DD29 pKa = 4.04DD30 pKa = 3.59IASVIISKK38 pKa = 10.64LEE40 pKa = 3.87PPEE43 pKa = 4.52VGLSTYY49 pKa = 10.74AKK51 pKa = 9.8RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84AKK56 pKa = 10.57KK57 pKa = 9.97LGRR60 pKa = 11.84CPRR63 pKa = 11.84CYY65 pKa = 9.88RR66 pKa = 11.84VIGLSQKK73 pKa = 10.51CDD75 pKa = 3.09GFKK78 pKa = 10.42CVPGISYY85 pKa = 9.83NRR87 pKa = 11.84KK88 pKa = 7.83VEE90 pKa = 3.86QNIRR94 pKa = 11.84YY95 pKa = 9.73GDD97 pKa = 3.32
Molecular weight: 11.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2743 |
67 |
1949 |
457.2 |
51.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.984 ± 0.441 | 2.771 ± 0.445 |
4.666 ± 0.44 | 7.437 ± 1.09 |
4.557 ± 0.348 | 5.724 ± 0.301 |
2.224 ± 0.243 | 4.521 ± 0.951 |
6.38 ± 0.505 | 9.807 ± 0.782 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.443 ± 0.361 | 3.5 ± 0.461 |
3.901 ± 0.967 | 2.661 ± 0.18 |
6.161 ± 0.594 | 7.838 ± 0.56 |
4.812 ± 0.469 | 8.057 ± 0.722 |
0.948 ± 0.136 | 3.609 ± 0.243 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
