
Alces alces faeces associated microvirus MP21 4718
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
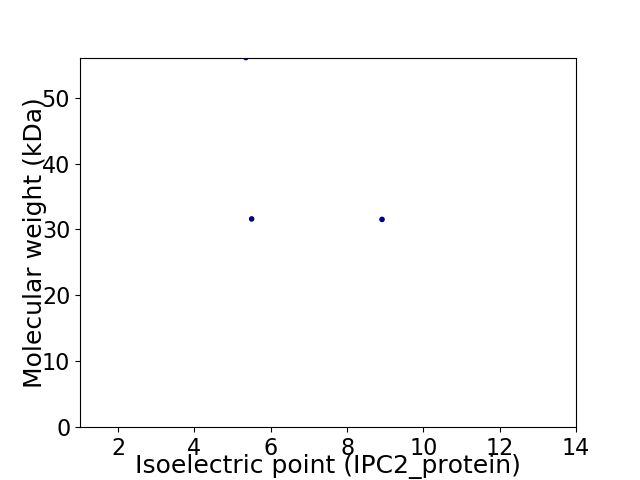
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CJ12|A0A2Z5CJ12_9VIRU Major capsid protein OS=Alces alces faeces associated microvirus MP21 4718 OX=2219138 PE=3 SV=1
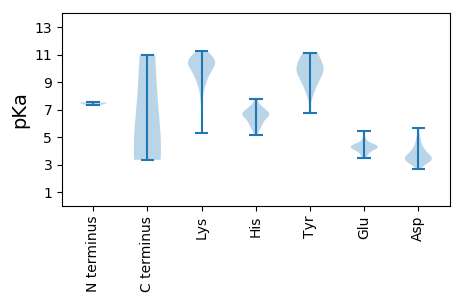
MM1 pKa = 7.47KK2 pKa = 10.22RR3 pKa = 11.84RR4 pKa = 11.84NKK6 pKa = 10.02FSLSHH11 pKa = 5.95YY12 pKa = 10.94VLGTCKK18 pKa = 9.46MGYY21 pKa = 8.12MLPIGLTEE29 pKa = 4.26VLPGDD34 pKa = 4.89TIQQATSVFMRR45 pKa = 11.84LAPMLAPVMHH55 pKa = 6.71PVQVKK60 pKa = 7.93IHH62 pKa = 5.79HH63 pKa = 6.6WYY65 pKa = 9.86VPLRR69 pKa = 11.84LVFPKK74 pKa = 10.62FEE76 pKa = 4.52DD77 pKa = 5.01FITGGPDD84 pKa = 3.33GLDD87 pKa = 3.01TTEE90 pKa = 4.87FPTITSDD97 pKa = 3.28EE98 pKa = 4.17TDD100 pKa = 3.26GFLEE104 pKa = 5.04GSLADD109 pKa = 4.31HH110 pKa = 7.04LGLPPGIPGLTVSALPFRR128 pKa = 11.84AYY130 pKa = 11.12NLIYY134 pKa = 10.53NEE136 pKa = 4.15WYY138 pKa = 10.0RR139 pKa = 11.84DD140 pKa = 3.15QDD142 pKa = 3.96LEE144 pKa = 4.32QEE146 pKa = 4.31IAISDD151 pKa = 3.8EE152 pKa = 4.35PGADD156 pKa = 2.9TTTSIEE162 pKa = 4.21LLRR165 pKa = 11.84GDD167 pKa = 3.53WEE169 pKa = 4.0RR170 pKa = 11.84DD171 pKa = 3.29YY172 pKa = 10.99FTTARR177 pKa = 11.84PWPQKK182 pKa = 10.73GIDD185 pKa = 3.46VTVPVQASGTADD197 pKa = 3.35VNIDD201 pKa = 2.96ITGNGTPSFSKK212 pKa = 10.82VSGFASGALQTNSSDD227 pKa = 3.74TIVTFGASSSGVSGPTSLAWLNPALQASGTVTGLDD262 pKa = 3.17VGSVNINDD270 pKa = 3.46LRR272 pKa = 11.84EE273 pKa = 4.07AFALQRR279 pKa = 11.84FEE281 pKa = 4.25EE282 pKa = 4.21HH283 pKa = 6.19RR284 pKa = 11.84AMYY287 pKa = 9.64GSRR290 pKa = 11.84YY291 pKa = 9.33VEE293 pKa = 3.84YY294 pKa = 10.74LRR296 pKa = 11.84YY297 pKa = 10.0LGIKK301 pKa = 10.33ASDD304 pKa = 3.48ARR306 pKa = 11.84LQRR309 pKa = 11.84PEE311 pKa = 3.75YY312 pKa = 10.59LGGGKK317 pKa = 7.49QTIQFSEE324 pKa = 4.46VLQTAADD331 pKa = 3.72GDD333 pKa = 4.29NPVGTLRR340 pKa = 11.84GHH342 pKa = 7.31GIAAMRR348 pKa = 11.84SNRR351 pKa = 11.84YY352 pKa = 8.7RR353 pKa = 11.84RR354 pKa = 11.84FIEE357 pKa = 3.55EE358 pKa = 3.45HH359 pKa = 6.62GYY361 pKa = 10.13IISIMLIRR369 pKa = 11.84PKK371 pKa = 10.28SIYY374 pKa = 8.5QQGVPRR380 pKa = 11.84TWLRR384 pKa = 11.84RR385 pKa = 11.84VKK387 pKa = 10.61EE388 pKa = 4.3DD389 pKa = 3.16FWQQEE394 pKa = 4.18LQHH397 pKa = 6.83IGQQEE402 pKa = 4.43VLTQEE407 pKa = 4.33LFGAAEE413 pKa = 4.06PEE415 pKa = 4.49KK416 pKa = 11.1VFGYY420 pKa = 8.13QNRR423 pKa = 11.84YY424 pKa = 9.41DD425 pKa = 4.75DD426 pKa = 4.13YY427 pKa = 9.1RR428 pKa = 11.84QHH430 pKa = 6.56VSYY433 pKa = 11.02VAGEE437 pKa = 3.84FRR439 pKa = 11.84SILNYY444 pKa = 8.59WHH446 pKa = 7.75LARR449 pKa = 11.84EE450 pKa = 4.43FGNEE454 pKa = 3.6PVLNADD460 pKa = 5.4FISSVPTEE468 pKa = 4.48RR469 pKa = 11.84IFAAQEE475 pKa = 3.54NDD477 pKa = 3.16QIYY480 pKa = 10.85YY481 pKa = 8.12MANHH485 pKa = 7.06SIQARR490 pKa = 11.84RR491 pKa = 11.84LMAKK495 pKa = 9.66YY496 pKa = 10.23GNPII500 pKa = 3.34
MM1 pKa = 7.47KK2 pKa = 10.22RR3 pKa = 11.84RR4 pKa = 11.84NKK6 pKa = 10.02FSLSHH11 pKa = 5.95YY12 pKa = 10.94VLGTCKK18 pKa = 9.46MGYY21 pKa = 8.12MLPIGLTEE29 pKa = 4.26VLPGDD34 pKa = 4.89TIQQATSVFMRR45 pKa = 11.84LAPMLAPVMHH55 pKa = 6.71PVQVKK60 pKa = 7.93IHH62 pKa = 5.79HH63 pKa = 6.6WYY65 pKa = 9.86VPLRR69 pKa = 11.84LVFPKK74 pKa = 10.62FEE76 pKa = 4.52DD77 pKa = 5.01FITGGPDD84 pKa = 3.33GLDD87 pKa = 3.01TTEE90 pKa = 4.87FPTITSDD97 pKa = 3.28EE98 pKa = 4.17TDD100 pKa = 3.26GFLEE104 pKa = 5.04GSLADD109 pKa = 4.31HH110 pKa = 7.04LGLPPGIPGLTVSALPFRR128 pKa = 11.84AYY130 pKa = 11.12NLIYY134 pKa = 10.53NEE136 pKa = 4.15WYY138 pKa = 10.0RR139 pKa = 11.84DD140 pKa = 3.15QDD142 pKa = 3.96LEE144 pKa = 4.32QEE146 pKa = 4.31IAISDD151 pKa = 3.8EE152 pKa = 4.35PGADD156 pKa = 2.9TTTSIEE162 pKa = 4.21LLRR165 pKa = 11.84GDD167 pKa = 3.53WEE169 pKa = 4.0RR170 pKa = 11.84DD171 pKa = 3.29YY172 pKa = 10.99FTTARR177 pKa = 11.84PWPQKK182 pKa = 10.73GIDD185 pKa = 3.46VTVPVQASGTADD197 pKa = 3.35VNIDD201 pKa = 2.96ITGNGTPSFSKK212 pKa = 10.82VSGFASGALQTNSSDD227 pKa = 3.74TIVTFGASSSGVSGPTSLAWLNPALQASGTVTGLDD262 pKa = 3.17VGSVNINDD270 pKa = 3.46LRR272 pKa = 11.84EE273 pKa = 4.07AFALQRR279 pKa = 11.84FEE281 pKa = 4.25EE282 pKa = 4.21HH283 pKa = 6.19RR284 pKa = 11.84AMYY287 pKa = 9.64GSRR290 pKa = 11.84YY291 pKa = 9.33VEE293 pKa = 3.84YY294 pKa = 10.74LRR296 pKa = 11.84YY297 pKa = 10.0LGIKK301 pKa = 10.33ASDD304 pKa = 3.48ARR306 pKa = 11.84LQRR309 pKa = 11.84PEE311 pKa = 3.75YY312 pKa = 10.59LGGGKK317 pKa = 7.49QTIQFSEE324 pKa = 4.46VLQTAADD331 pKa = 3.72GDD333 pKa = 4.29NPVGTLRR340 pKa = 11.84GHH342 pKa = 7.31GIAAMRR348 pKa = 11.84SNRR351 pKa = 11.84YY352 pKa = 8.7RR353 pKa = 11.84RR354 pKa = 11.84FIEE357 pKa = 3.55EE358 pKa = 3.45HH359 pKa = 6.62GYY361 pKa = 10.13IISIMLIRR369 pKa = 11.84PKK371 pKa = 10.28SIYY374 pKa = 8.5QQGVPRR380 pKa = 11.84TWLRR384 pKa = 11.84RR385 pKa = 11.84VKK387 pKa = 10.61EE388 pKa = 4.3DD389 pKa = 3.16FWQQEE394 pKa = 4.18LQHH397 pKa = 6.83IGQQEE402 pKa = 4.43VLTQEE407 pKa = 4.33LFGAAEE413 pKa = 4.06PEE415 pKa = 4.49KK416 pKa = 11.1VFGYY420 pKa = 8.13QNRR423 pKa = 11.84YY424 pKa = 9.41DD425 pKa = 4.75DD426 pKa = 4.13YY427 pKa = 9.1RR428 pKa = 11.84QHH430 pKa = 6.56VSYY433 pKa = 11.02VAGEE437 pKa = 3.84FRR439 pKa = 11.84SILNYY444 pKa = 8.59WHH446 pKa = 7.75LARR449 pKa = 11.84EE450 pKa = 4.43FGNEE454 pKa = 3.6PVLNADD460 pKa = 5.4FISSVPTEE468 pKa = 4.48RR469 pKa = 11.84IFAAQEE475 pKa = 3.54NDD477 pKa = 3.16QIYY480 pKa = 10.85YY481 pKa = 8.12MANHH485 pKa = 7.06SIQARR490 pKa = 11.84RR491 pKa = 11.84LMAKK495 pKa = 9.66YY496 pKa = 10.23GNPII500 pKa = 3.34
Molecular weight: 56.07 kDa
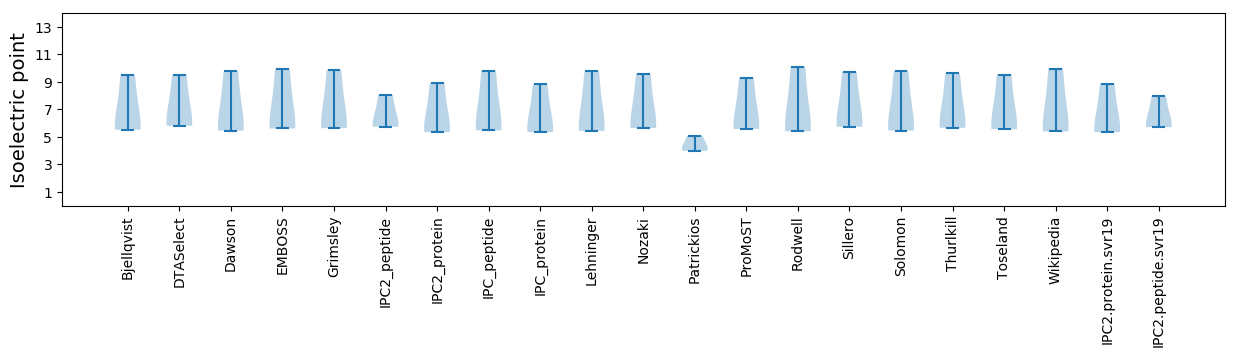
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CJ12|A0A2Z5CJ12_9VIRU Major capsid protein OS=Alces alces faeces associated microvirus MP21 4718 OX=2219138 PE=3 SV=1
MM1 pKa = 7.51LEE3 pKa = 4.1SLEE6 pKa = 4.47HH7 pKa = 6.55EE8 pKa = 4.17EE9 pKa = 4.87SSFITLTYY17 pKa = 10.06KK18 pKa = 10.86EE19 pKa = 4.57DD20 pKa = 3.65MLPFMVDD27 pKa = 3.44LPVPTLCVRR36 pKa = 11.84DD37 pKa = 3.42AQLFLKK43 pKa = 10.35RR44 pKa = 11.84LRR46 pKa = 11.84KK47 pKa = 9.99GLTDD51 pKa = 3.56HH52 pKa = 6.8ARR54 pKa = 11.84KK55 pKa = 9.53EE56 pKa = 4.36IKK58 pKa = 10.33KK59 pKa = 8.66KK60 pKa = 8.81TGKK63 pKa = 10.19KK64 pKa = 9.56KK65 pKa = 10.72LKK67 pKa = 10.4LPSFQFRR74 pKa = 11.84YY75 pKa = 9.13YY76 pKa = 10.94LCGEE80 pKa = 3.87YY81 pKa = 10.46GPRR84 pKa = 11.84GGRR87 pKa = 11.84PHH89 pKa = 5.63YY90 pKa = 10.04HH91 pKa = 7.02AILFGVPPDD100 pKa = 3.3AAKK103 pKa = 10.48FIEE106 pKa = 4.53KK107 pKa = 9.76AWSIEE112 pKa = 3.67GEE114 pKa = 4.16PIGRR118 pKa = 11.84IDD120 pKa = 3.48VQPINEE126 pKa = 4.0NNIQYY131 pKa = 9.85VAGYY135 pKa = 6.78VTKK138 pKa = 10.89KK139 pKa = 5.31MTRR142 pKa = 11.84RR143 pKa = 11.84GDD145 pKa = 3.15GRR147 pKa = 11.84KK148 pKa = 9.36PEE150 pKa = 3.98FSIMSRR156 pKa = 11.84KK157 pKa = 9.41PGIGANAVPKK167 pKa = 10.45LVEE170 pKa = 4.46LFSNPDD176 pKa = 2.86TGQYY180 pKa = 10.49FDD182 pKa = 4.66IVHH185 pKa = 6.47EE186 pKa = 4.74APVLMHH192 pKa = 6.61GRR194 pKa = 11.84KK195 pKa = 8.84QLPLGRR201 pKa = 11.84YY202 pKa = 9.1LRR204 pKa = 11.84DD205 pKa = 3.39KK206 pKa = 11.26VLDD209 pKa = 3.76EE210 pKa = 4.62MGLEE214 pKa = 4.23RR215 pKa = 11.84DD216 pKa = 3.39LTPYY220 pKa = 10.01IEE222 pKa = 5.43EE223 pKa = 4.13MFQKK227 pKa = 10.6YY228 pKa = 9.91LEE230 pKa = 4.32AKK232 pKa = 10.3RR233 pKa = 11.84SNKK236 pKa = 9.37YY237 pKa = 8.83VNCYY241 pKa = 9.85LEE243 pKa = 4.68RR244 pKa = 11.84LLIDD248 pKa = 3.42EE249 pKa = 4.42SKK251 pKa = 10.7QRR253 pKa = 11.84NLQMRR258 pKa = 11.84AIHH261 pKa = 6.72KK262 pKa = 9.93IYY264 pKa = 10.53NSRR267 pKa = 11.84NKK269 pKa = 10.55AII271 pKa = 4.02
MM1 pKa = 7.51LEE3 pKa = 4.1SLEE6 pKa = 4.47HH7 pKa = 6.55EE8 pKa = 4.17EE9 pKa = 4.87SSFITLTYY17 pKa = 10.06KK18 pKa = 10.86EE19 pKa = 4.57DD20 pKa = 3.65MLPFMVDD27 pKa = 3.44LPVPTLCVRR36 pKa = 11.84DD37 pKa = 3.42AQLFLKK43 pKa = 10.35RR44 pKa = 11.84LRR46 pKa = 11.84KK47 pKa = 9.99GLTDD51 pKa = 3.56HH52 pKa = 6.8ARR54 pKa = 11.84KK55 pKa = 9.53EE56 pKa = 4.36IKK58 pKa = 10.33KK59 pKa = 8.66KK60 pKa = 8.81TGKK63 pKa = 10.19KK64 pKa = 9.56KK65 pKa = 10.72LKK67 pKa = 10.4LPSFQFRR74 pKa = 11.84YY75 pKa = 9.13YY76 pKa = 10.94LCGEE80 pKa = 3.87YY81 pKa = 10.46GPRR84 pKa = 11.84GGRR87 pKa = 11.84PHH89 pKa = 5.63YY90 pKa = 10.04HH91 pKa = 7.02AILFGVPPDD100 pKa = 3.3AAKK103 pKa = 10.48FIEE106 pKa = 4.53KK107 pKa = 9.76AWSIEE112 pKa = 3.67GEE114 pKa = 4.16PIGRR118 pKa = 11.84IDD120 pKa = 3.48VQPINEE126 pKa = 4.0NNIQYY131 pKa = 9.85VAGYY135 pKa = 6.78VTKK138 pKa = 10.89KK139 pKa = 5.31MTRR142 pKa = 11.84RR143 pKa = 11.84GDD145 pKa = 3.15GRR147 pKa = 11.84KK148 pKa = 9.36PEE150 pKa = 3.98FSIMSRR156 pKa = 11.84KK157 pKa = 9.41PGIGANAVPKK167 pKa = 10.45LVEE170 pKa = 4.46LFSNPDD176 pKa = 2.86TGQYY180 pKa = 10.49FDD182 pKa = 4.66IVHH185 pKa = 6.47EE186 pKa = 4.74APVLMHH192 pKa = 6.61GRR194 pKa = 11.84KK195 pKa = 8.84QLPLGRR201 pKa = 11.84YY202 pKa = 9.1LRR204 pKa = 11.84DD205 pKa = 3.39KK206 pKa = 11.26VLDD209 pKa = 3.76EE210 pKa = 4.62MGLEE214 pKa = 4.23RR215 pKa = 11.84DD216 pKa = 3.39LTPYY220 pKa = 10.01IEE222 pKa = 5.43EE223 pKa = 4.13MFQKK227 pKa = 10.6YY228 pKa = 9.91LEE230 pKa = 4.32AKK232 pKa = 10.3RR233 pKa = 11.84SNKK236 pKa = 9.37YY237 pKa = 8.83VNCYY241 pKa = 9.85LEE243 pKa = 4.68RR244 pKa = 11.84LLIDD248 pKa = 3.42EE249 pKa = 4.42SKK251 pKa = 10.7QRR253 pKa = 11.84NLQMRR258 pKa = 11.84AIHH261 pKa = 6.72KK262 pKa = 9.93IYY264 pKa = 10.53NSRR267 pKa = 11.84NKK269 pKa = 10.55AII271 pKa = 4.02
Molecular weight: 31.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1064 |
271 |
500 |
354.7 |
39.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.613 ± 1.387 | 0.376 ± 0.274 |
5.451 ± 0.303 | 6.109 ± 0.674 |
3.853 ± 0.568 | 8.835 ± 1.042 |
2.162 ± 0.303 | 5.545 ± 0.673 |
4.511 ± 1.893 | 8.271 ± 0.941 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.726 ± 0.32 | 3.853 ± 0.22 |
5.451 ± 0.418 | 5.451 ± 0.94 |
6.767 ± 0.366 | 6.955 ± 1.395 |
4.887 ± 0.908 | 5.357 ± 0.382 |
1.222 ± 0.324 | 4.605 ± 0.241 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
