
Molossus molossus circovirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 7.79
Get precalculated fractions of proteins
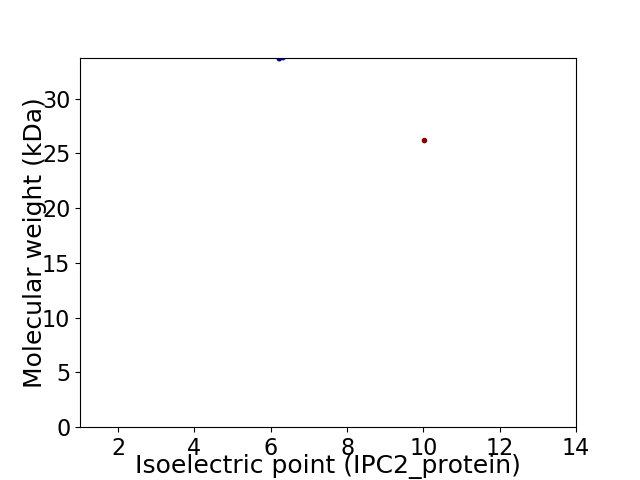
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
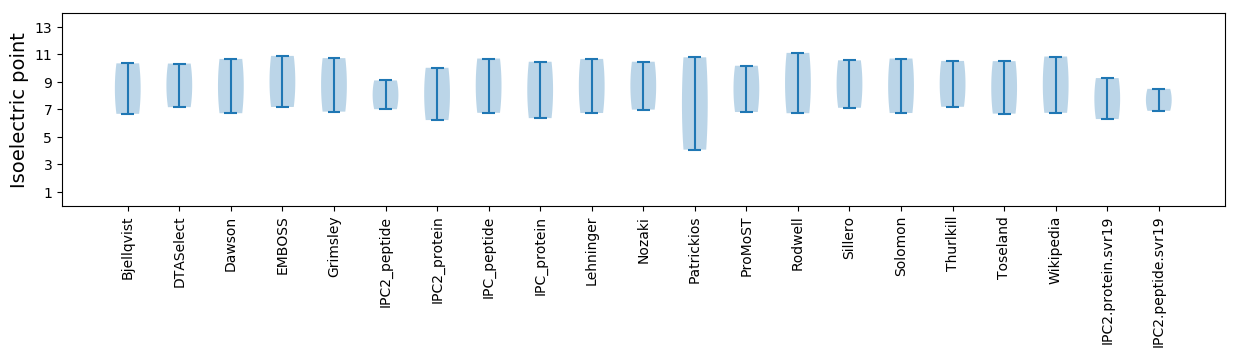
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I2MP65|A0A2I2MP65_9CIRC Capsid OS=Molossus molossus circovirus 3 OX=1959844 GN=Cap PE=4 SV=1
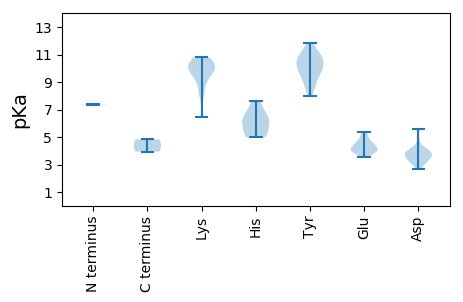
MM1 pKa = 7.39QSRR4 pKa = 11.84HH5 pKa = 5.0YY6 pKa = 10.78CFTLNNPKK14 pKa = 10.28EE15 pKa = 4.65LIDD18 pKa = 3.88FTTIPHH24 pKa = 5.06VRR26 pKa = 11.84YY27 pKa = 9.79AIYY30 pKa = 10.12QEE32 pKa = 4.66EE33 pKa = 4.25IGEE36 pKa = 4.24SGNYY40 pKa = 8.94HH41 pKa = 5.03FQGYY45 pKa = 9.72IEE47 pKa = 4.5FDD49 pKa = 3.07RR50 pKa = 11.84PTRR53 pKa = 11.84LGFLQRR59 pKa = 11.84HH60 pKa = 5.7LNGGHH65 pKa = 6.03FEE67 pKa = 4.1LRR69 pKa = 11.84RR70 pKa = 11.84GSRR73 pKa = 11.84DD74 pKa = 3.0EE75 pKa = 3.74ARR77 pKa = 11.84EE78 pKa = 3.81YY79 pKa = 10.66CRR81 pKa = 11.84KK82 pKa = 9.48LDD84 pKa = 3.64SRR86 pKa = 11.84IGEE89 pKa = 4.14VFEE92 pKa = 5.08YY93 pKa = 11.07GDD95 pKa = 4.0WDD97 pKa = 3.29QGGQGVRR104 pKa = 11.84TDD106 pKa = 4.04FNALHH111 pKa = 6.07EE112 pKa = 4.65LVRR115 pKa = 11.84QGTPTSEE122 pKa = 5.15IMDD125 pKa = 3.51QMPSMFYY132 pKa = 9.96RR133 pKa = 11.84YY134 pKa = 9.66HH135 pKa = 7.65SGIGKK140 pKa = 9.02AQLLCVQDD148 pKa = 4.17RR149 pKa = 11.84DD150 pKa = 3.71WPTKK154 pKa = 10.34IIVLYY159 pKa = 9.97GASGTGKK166 pKa = 10.2SRR168 pKa = 11.84WAHH171 pKa = 6.41DD172 pKa = 3.72YY173 pKa = 10.31CHH175 pKa = 7.23PSHH178 pKa = 6.51QYY180 pKa = 8.55WVSNTKK186 pKa = 9.54WFDD189 pKa = 3.3GYY191 pKa = 10.77EE192 pKa = 4.01FQPVVVLDD200 pKa = 4.77DD201 pKa = 3.86FRR203 pKa = 11.84GWLPLHH209 pKa = 5.84QLLRR213 pKa = 11.84LCDD216 pKa = 3.96RR217 pKa = 11.84YY218 pKa = 10.41PYY220 pKa = 8.73TVEE223 pKa = 3.51WKK225 pKa = 9.88GGSRR229 pKa = 11.84KK230 pKa = 8.16FTAHH234 pKa = 6.34VIIITSTRR242 pKa = 11.84RR243 pKa = 11.84PDD245 pKa = 2.7EE246 pKa = 4.04WYY248 pKa = 9.92NWEE251 pKa = 4.53EE252 pKa = 3.9LHH254 pKa = 7.05EE255 pKa = 5.37DD256 pKa = 3.26PAQLFRR262 pKa = 11.84RR263 pKa = 11.84IDD265 pKa = 3.5EE266 pKa = 4.37FLGRR270 pKa = 11.84TPRR273 pKa = 11.84RR274 pKa = 11.84SRR276 pKa = 11.84TALSEE281 pKa = 4.05DD282 pKa = 3.48RR283 pKa = 4.83
MM1 pKa = 7.39QSRR4 pKa = 11.84HH5 pKa = 5.0YY6 pKa = 10.78CFTLNNPKK14 pKa = 10.28EE15 pKa = 4.65LIDD18 pKa = 3.88FTTIPHH24 pKa = 5.06VRR26 pKa = 11.84YY27 pKa = 9.79AIYY30 pKa = 10.12QEE32 pKa = 4.66EE33 pKa = 4.25IGEE36 pKa = 4.24SGNYY40 pKa = 8.94HH41 pKa = 5.03FQGYY45 pKa = 9.72IEE47 pKa = 4.5FDD49 pKa = 3.07RR50 pKa = 11.84PTRR53 pKa = 11.84LGFLQRR59 pKa = 11.84HH60 pKa = 5.7LNGGHH65 pKa = 6.03FEE67 pKa = 4.1LRR69 pKa = 11.84RR70 pKa = 11.84GSRR73 pKa = 11.84DD74 pKa = 3.0EE75 pKa = 3.74ARR77 pKa = 11.84EE78 pKa = 3.81YY79 pKa = 10.66CRR81 pKa = 11.84KK82 pKa = 9.48LDD84 pKa = 3.64SRR86 pKa = 11.84IGEE89 pKa = 4.14VFEE92 pKa = 5.08YY93 pKa = 11.07GDD95 pKa = 4.0WDD97 pKa = 3.29QGGQGVRR104 pKa = 11.84TDD106 pKa = 4.04FNALHH111 pKa = 6.07EE112 pKa = 4.65LVRR115 pKa = 11.84QGTPTSEE122 pKa = 5.15IMDD125 pKa = 3.51QMPSMFYY132 pKa = 9.96RR133 pKa = 11.84YY134 pKa = 9.66HH135 pKa = 7.65SGIGKK140 pKa = 9.02AQLLCVQDD148 pKa = 4.17RR149 pKa = 11.84DD150 pKa = 3.71WPTKK154 pKa = 10.34IIVLYY159 pKa = 9.97GASGTGKK166 pKa = 10.2SRR168 pKa = 11.84WAHH171 pKa = 6.41DD172 pKa = 3.72YY173 pKa = 10.31CHH175 pKa = 7.23PSHH178 pKa = 6.51QYY180 pKa = 8.55WVSNTKK186 pKa = 9.54WFDD189 pKa = 3.3GYY191 pKa = 10.77EE192 pKa = 4.01FQPVVVLDD200 pKa = 4.77DD201 pKa = 3.86FRR203 pKa = 11.84GWLPLHH209 pKa = 5.84QLLRR213 pKa = 11.84LCDD216 pKa = 3.96RR217 pKa = 11.84YY218 pKa = 10.41PYY220 pKa = 8.73TVEE223 pKa = 3.51WKK225 pKa = 9.88GGSRR229 pKa = 11.84KK230 pKa = 8.16FTAHH234 pKa = 6.34VIIITSTRR242 pKa = 11.84RR243 pKa = 11.84PDD245 pKa = 2.7EE246 pKa = 4.04WYY248 pKa = 9.92NWEE251 pKa = 4.53EE252 pKa = 3.9LHH254 pKa = 7.05EE255 pKa = 5.37DD256 pKa = 3.26PAQLFRR262 pKa = 11.84RR263 pKa = 11.84IDD265 pKa = 3.5EE266 pKa = 4.37FLGRR270 pKa = 11.84TPRR273 pKa = 11.84RR274 pKa = 11.84SRR276 pKa = 11.84TALSEE281 pKa = 4.05DD282 pKa = 3.48RR283 pKa = 4.83
Molecular weight: 33.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I2MP65|A0A2I2MP65_9CIRC Capsid OS=Molossus molossus circovirus 3 OX=1959844 GN=Cap PE=4 SV=1
MM1 pKa = 7.36RR2 pKa = 11.84RR3 pKa = 11.84KK4 pKa = 9.39FRR6 pKa = 11.84RR7 pKa = 11.84FRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 8.41FKK13 pKa = 10.5KK14 pKa = 9.62FSRR17 pKa = 11.84RR18 pKa = 11.84FKK20 pKa = 10.22RR21 pKa = 11.84HH22 pKa = 5.26FGGKK26 pKa = 8.2RR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 6.46TTRR32 pKa = 11.84QVQFKK37 pKa = 9.91FKK39 pKa = 10.15VQTVPYY45 pKa = 9.11LNGSIAPSSSINWNNTSNTASHH67 pKa = 5.4YY68 pKa = 9.69TFAFTLGDD76 pKa = 3.45IPHH79 pKa = 6.55YY80 pKa = 10.65SDD82 pKa = 4.11LSSVFDD88 pKa = 3.9AAKK91 pKa = 10.18LAAVKK96 pKa = 10.46LKK98 pKa = 9.77FVPRR102 pKa = 11.84YY103 pKa = 8.81TMGQLPTSASTTYY116 pKa = 11.31ANTSTPCVVVKK127 pKa = 10.53DD128 pKa = 4.05YY129 pKa = 11.81DD130 pKa = 3.78DD131 pKa = 5.57ANPLTSYY138 pKa = 11.67ANALLYY144 pKa = 10.79QNARR148 pKa = 11.84VVSILKK154 pKa = 9.83PFSVYY159 pKa = 10.8LKK161 pKa = 9.24PKK163 pKa = 10.19LSGGVEE169 pKa = 3.98NTSLVIVAQSQARR182 pKa = 11.84PWLDD186 pKa = 3.1SGATAVPYY194 pKa = 10.56YY195 pKa = 10.25GVKK198 pKa = 10.64LEE200 pKa = 4.12VPGINTTQMLGQAIWDD216 pKa = 3.6IYY218 pKa = 7.95GTYY221 pKa = 9.42YY222 pKa = 11.11VKK224 pKa = 10.81LKK226 pKa = 10.36QIRR229 pKa = 11.84LLL231 pKa = 3.93
MM1 pKa = 7.36RR2 pKa = 11.84RR3 pKa = 11.84KK4 pKa = 9.39FRR6 pKa = 11.84RR7 pKa = 11.84FRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 8.41FKK13 pKa = 10.5KK14 pKa = 9.62FSRR17 pKa = 11.84RR18 pKa = 11.84FKK20 pKa = 10.22RR21 pKa = 11.84HH22 pKa = 5.26FGGKK26 pKa = 8.2RR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 6.46TTRR32 pKa = 11.84QVQFKK37 pKa = 9.91FKK39 pKa = 10.15VQTVPYY45 pKa = 9.11LNGSIAPSSSINWNNTSNTASHH67 pKa = 5.4YY68 pKa = 9.69TFAFTLGDD76 pKa = 3.45IPHH79 pKa = 6.55YY80 pKa = 10.65SDD82 pKa = 4.11LSSVFDD88 pKa = 3.9AAKK91 pKa = 10.18LAAVKK96 pKa = 10.46LKK98 pKa = 9.77FVPRR102 pKa = 11.84YY103 pKa = 8.81TMGQLPTSASTTYY116 pKa = 11.31ANTSTPCVVVKK127 pKa = 10.53DD128 pKa = 4.05YY129 pKa = 11.81DD130 pKa = 3.78DD131 pKa = 5.57ANPLTSYY138 pKa = 11.67ANALLYY144 pKa = 10.79QNARR148 pKa = 11.84VVSILKK154 pKa = 9.83PFSVYY159 pKa = 10.8LKK161 pKa = 9.24PKK163 pKa = 10.19LSGGVEE169 pKa = 3.98NTSLVIVAQSQARR182 pKa = 11.84PWLDD186 pKa = 3.1SGATAVPYY194 pKa = 10.56YY195 pKa = 10.25GVKK198 pKa = 10.64LEE200 pKa = 4.12VPGINTTQMLGQAIWDD216 pKa = 3.6IYY218 pKa = 7.95GTYY221 pKa = 9.42YY222 pKa = 11.11VKK224 pKa = 10.81LKK226 pKa = 10.36QIRR229 pKa = 11.84LLL231 pKa = 3.93
Molecular weight: 26.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
514 |
231 |
283 |
257.0 |
29.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.253 ± 1.516 | 1.167 ± 0.438 |
5.253 ± 1.068 | 4.28 ± 2.038 |
5.447 ± 0.108 | 6.809 ± 0.964 |
3.113 ± 1.083 | 4.475 ± 0.345 |
5.253 ± 1.774 | 7.977 ± 0.148 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.362 ± 0.038 | 3.502 ± 0.752 |
4.864 ± 0.198 | 4.669 ± 0.203 |
8.755 ± 1.092 | 6.809 ± 1.104 |
6.809 ± 0.845 | 6.031 ± 1.31 |
2.335 ± 0.618 | 5.837 ± 0.134 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
