
Rice stripe mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus; Rice stripe mosaic cytorhabdovirus
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
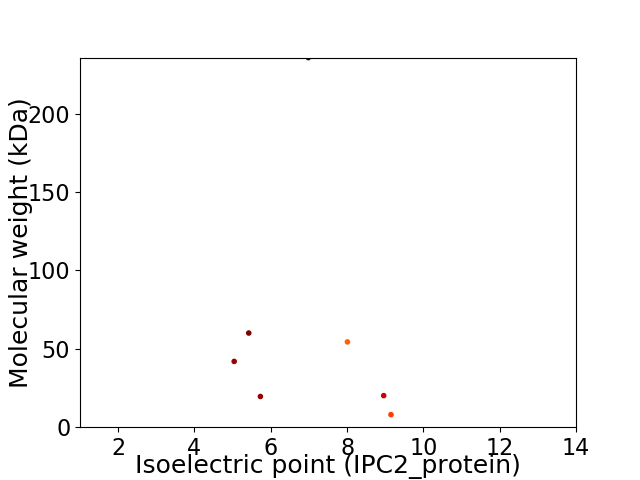
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1P8D6S4|A0A1P8D6S4_9RHAB Nucleocapsid protein OS=Rice stripe mosaic virus OX=1931356 PE=4 SV=1
MM1 pKa = 7.46SVPEE5 pKa = 4.09DD6 pKa = 3.84TPFRR10 pKa = 11.84SYY12 pKa = 11.63SSIFDD17 pKa = 4.04DD18 pKa = 4.2SDD20 pKa = 3.69FVQPQPMSFKK30 pKa = 9.59ATKK33 pKa = 9.8EE34 pKa = 4.17SEE36 pKa = 4.37SLPEE40 pKa = 4.01TEE42 pKa = 4.98KK43 pKa = 11.28EE44 pKa = 4.21DD45 pKa = 3.38MSTEE49 pKa = 4.05YY50 pKa = 10.74LSEE53 pKa = 4.07PLRR56 pKa = 11.84TKK58 pKa = 10.31SGKK61 pKa = 9.53KK62 pKa = 8.14NRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 10.45GKK69 pKa = 9.21DD70 pKa = 2.95LKK72 pKa = 11.39SLFTQEE78 pKa = 4.41AGLPAPEE85 pKa = 5.01ADD87 pKa = 3.37SVLPEE92 pKa = 3.95SSPYY96 pKa = 10.95EE97 pKa = 3.95NDD99 pKa = 3.15NAQLEE104 pKa = 4.5LPKK107 pKa = 10.41PILKK111 pKa = 9.66TSDD114 pKa = 3.1APVFLRR120 pKa = 11.84EE121 pKa = 4.01KK122 pKa = 10.69DD123 pKa = 3.5LSKK126 pKa = 11.11EE127 pKa = 3.48FAAACKK133 pKa = 10.03TNGILPRR140 pKa = 11.84DD141 pKa = 3.53EE142 pKa = 4.43WKK144 pKa = 10.92SSVAAKK150 pKa = 9.57YY151 pKa = 9.65HH152 pKa = 5.7AEE154 pKa = 3.97EE155 pKa = 4.43GKK157 pKa = 7.84MTKK160 pKa = 9.76RR161 pKa = 11.84DD162 pKa = 2.87ISLIVFGMEE171 pKa = 3.9LYY173 pKa = 10.37KK174 pKa = 10.42RR175 pKa = 11.84YY176 pKa = 9.82NVEE179 pKa = 4.27SEE181 pKa = 4.03VSTLFTSLVTEE192 pKa = 4.12LQGIKK197 pKa = 10.12VAAKK201 pKa = 9.15EE202 pKa = 4.02LNDD205 pKa = 3.5TRR207 pKa = 11.84EE208 pKa = 4.27VLTKK212 pKa = 10.08IPGEE216 pKa = 3.76IVSAVKK222 pKa = 10.61AGVKK226 pKa = 9.86EE227 pKa = 4.11GTEE230 pKa = 3.64MGMDD234 pKa = 3.94YY235 pKa = 10.82IEE237 pKa = 4.6TRR239 pKa = 11.84TKK241 pKa = 10.38VAPKK245 pKa = 9.91SAPKK249 pKa = 10.32VDD251 pKa = 3.08ISKK254 pKa = 10.31PMSSKK259 pKa = 10.28MMEE262 pKa = 4.16QQDD265 pKa = 3.77EE266 pKa = 4.5SSDD269 pKa = 3.5EE270 pKa = 4.26SSDD273 pKa = 3.77NEE275 pKa = 4.19SEE277 pKa = 4.14EE278 pKa = 4.29SEE280 pKa = 4.25EE281 pKa = 4.3EE282 pKa = 4.19SFEE285 pKa = 4.02TKK287 pKa = 10.39AAIFLALIKK296 pKa = 10.73VPEE299 pKa = 4.17EE300 pKa = 3.8EE301 pKa = 3.89RR302 pKa = 11.84DD303 pKa = 3.61NAIVLMALRR312 pKa = 11.84AVISDD317 pKa = 3.91SEE319 pKa = 4.37LNQAIRR325 pKa = 11.84NDD327 pKa = 4.27RR328 pKa = 11.84ISSSVADD335 pKa = 4.2MYY337 pKa = 10.43HH338 pKa = 6.17QKK340 pKa = 10.32ISDD343 pKa = 3.7KK344 pKa = 10.98ARR346 pKa = 11.84EE347 pKa = 3.98LMGKK351 pKa = 10.29GKK353 pKa = 8.22TNKK356 pKa = 9.56RR357 pKa = 11.84AKK359 pKa = 9.6QPKK362 pKa = 7.39SSKK365 pKa = 10.03YY366 pKa = 10.53ASDD369 pKa = 4.44YY370 pKa = 11.43YY371 pKa = 11.44DD372 pKa = 4.1DD373 pKa = 4.87ALL375 pKa = 5.77
MM1 pKa = 7.46SVPEE5 pKa = 4.09DD6 pKa = 3.84TPFRR10 pKa = 11.84SYY12 pKa = 11.63SSIFDD17 pKa = 4.04DD18 pKa = 4.2SDD20 pKa = 3.69FVQPQPMSFKK30 pKa = 9.59ATKK33 pKa = 9.8EE34 pKa = 4.17SEE36 pKa = 4.37SLPEE40 pKa = 4.01TEE42 pKa = 4.98KK43 pKa = 11.28EE44 pKa = 4.21DD45 pKa = 3.38MSTEE49 pKa = 4.05YY50 pKa = 10.74LSEE53 pKa = 4.07PLRR56 pKa = 11.84TKK58 pKa = 10.31SGKK61 pKa = 9.53KK62 pKa = 8.14NRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 10.45GKK69 pKa = 9.21DD70 pKa = 2.95LKK72 pKa = 11.39SLFTQEE78 pKa = 4.41AGLPAPEE85 pKa = 5.01ADD87 pKa = 3.37SVLPEE92 pKa = 3.95SSPYY96 pKa = 10.95EE97 pKa = 3.95NDD99 pKa = 3.15NAQLEE104 pKa = 4.5LPKK107 pKa = 10.41PILKK111 pKa = 9.66TSDD114 pKa = 3.1APVFLRR120 pKa = 11.84EE121 pKa = 4.01KK122 pKa = 10.69DD123 pKa = 3.5LSKK126 pKa = 11.11EE127 pKa = 3.48FAAACKK133 pKa = 10.03TNGILPRR140 pKa = 11.84DD141 pKa = 3.53EE142 pKa = 4.43WKK144 pKa = 10.92SSVAAKK150 pKa = 9.57YY151 pKa = 9.65HH152 pKa = 5.7AEE154 pKa = 3.97EE155 pKa = 4.43GKK157 pKa = 7.84MTKK160 pKa = 9.76RR161 pKa = 11.84DD162 pKa = 2.87ISLIVFGMEE171 pKa = 3.9LYY173 pKa = 10.37KK174 pKa = 10.42RR175 pKa = 11.84YY176 pKa = 9.82NVEE179 pKa = 4.27SEE181 pKa = 4.03VSTLFTSLVTEE192 pKa = 4.12LQGIKK197 pKa = 10.12VAAKK201 pKa = 9.15EE202 pKa = 4.02LNDD205 pKa = 3.5TRR207 pKa = 11.84EE208 pKa = 4.27VLTKK212 pKa = 10.08IPGEE216 pKa = 3.76IVSAVKK222 pKa = 10.61AGVKK226 pKa = 9.86EE227 pKa = 4.11GTEE230 pKa = 3.64MGMDD234 pKa = 3.94YY235 pKa = 10.82IEE237 pKa = 4.6TRR239 pKa = 11.84TKK241 pKa = 10.38VAPKK245 pKa = 9.91SAPKK249 pKa = 10.32VDD251 pKa = 3.08ISKK254 pKa = 10.31PMSSKK259 pKa = 10.28MMEE262 pKa = 4.16QQDD265 pKa = 3.77EE266 pKa = 4.5SSDD269 pKa = 3.5EE270 pKa = 4.26SSDD273 pKa = 3.77NEE275 pKa = 4.19SEE277 pKa = 4.14EE278 pKa = 4.29SEE280 pKa = 4.25EE281 pKa = 4.3EE282 pKa = 4.19SFEE285 pKa = 4.02TKK287 pKa = 10.39AAIFLALIKK296 pKa = 10.73VPEE299 pKa = 4.17EE300 pKa = 3.8EE301 pKa = 3.89RR302 pKa = 11.84DD303 pKa = 3.61NAIVLMALRR312 pKa = 11.84AVISDD317 pKa = 3.91SEE319 pKa = 4.37LNQAIRR325 pKa = 11.84NDD327 pKa = 4.27RR328 pKa = 11.84ISSSVADD335 pKa = 4.2MYY337 pKa = 10.43HH338 pKa = 6.17QKK340 pKa = 10.32ISDD343 pKa = 3.7KK344 pKa = 10.98ARR346 pKa = 11.84EE347 pKa = 3.98LMGKK351 pKa = 10.29GKK353 pKa = 8.22TNKK356 pKa = 9.56RR357 pKa = 11.84AKK359 pKa = 9.6QPKK362 pKa = 7.39SSKK365 pKa = 10.03YY366 pKa = 10.53ASDD369 pKa = 4.44YY370 pKa = 11.43YY371 pKa = 11.44DD372 pKa = 4.1DD373 pKa = 4.87ALL375 pKa = 5.77
Molecular weight: 41.97 kDa
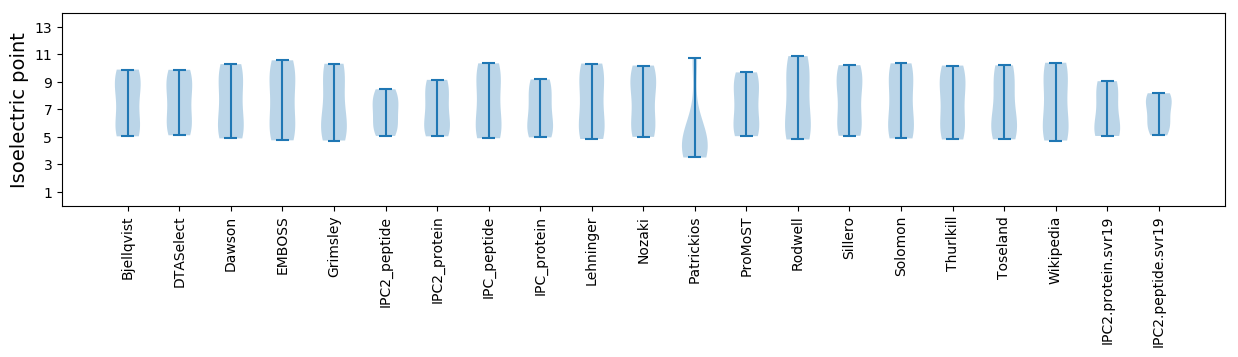
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8D6S6|A0A1P8D6S6_9RHAB Matrix protein OS=Rice stripe mosaic virus OX=1931356 PE=4 SV=1
MM1 pKa = 7.44EE2 pKa = 5.86FNWPWGQNSEE12 pKa = 4.37TEE14 pKa = 3.71ITKK17 pKa = 10.16NLRR20 pKa = 11.84FEE22 pKa = 5.33DD23 pKa = 3.8IKK25 pKa = 11.67VMAIIILVWVKK36 pKa = 10.72CLLIYY41 pKa = 10.29HH42 pKa = 6.92FKK44 pKa = 10.77RR45 pKa = 11.84KK46 pKa = 8.95IRR48 pKa = 11.84RR49 pKa = 11.84LRR51 pKa = 11.84SLLIKK56 pKa = 10.7GSSQWVLHH64 pKa = 6.44DD65 pKa = 3.73AA66 pKa = 4.44
MM1 pKa = 7.44EE2 pKa = 5.86FNWPWGQNSEE12 pKa = 4.37TEE14 pKa = 3.71ITKK17 pKa = 10.16NLRR20 pKa = 11.84FEE22 pKa = 5.33DD23 pKa = 3.8IKK25 pKa = 11.67VMAIIILVWVKK36 pKa = 10.72CLLIYY41 pKa = 10.29HH42 pKa = 6.92FKK44 pKa = 10.77RR45 pKa = 11.84KK46 pKa = 8.95IRR48 pKa = 11.84RR49 pKa = 11.84LRR51 pKa = 11.84SLLIKK56 pKa = 10.7GSSQWVLHH64 pKa = 6.44DD65 pKa = 3.73AA66 pKa = 4.44
Molecular weight: 8.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3885 |
66 |
2066 |
555.0 |
62.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.255 ± 1.298 | 1.57 ± 0.459 |
5.663 ± 0.393 | 6.795 ± 0.905 |
3.604 ± 0.198 | 5.122 ± 0.471 |
1.673 ± 0.433 | 6.461 ± 0.738 |
7.027 ± 1.47 | 9.704 ± 1.001 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.192 ± 0.224 | 3.629 ± 0.132 |
4.633 ± 0.39 | 2.754 ± 0.169 |
5.431 ± 0.762 | 8.623 ± 0.84 |
6.435 ± 0.735 | 5.92 ± 0.467 |
1.493 ± 0.339 | 4.015 ± 0.245 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
