
Chinook salmon bafinivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Tornidovirineae; Tobaniviridae; Piscanivirinae; Oncotshavirus; Salnivirus; Chinook salmon nidovirus 1
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
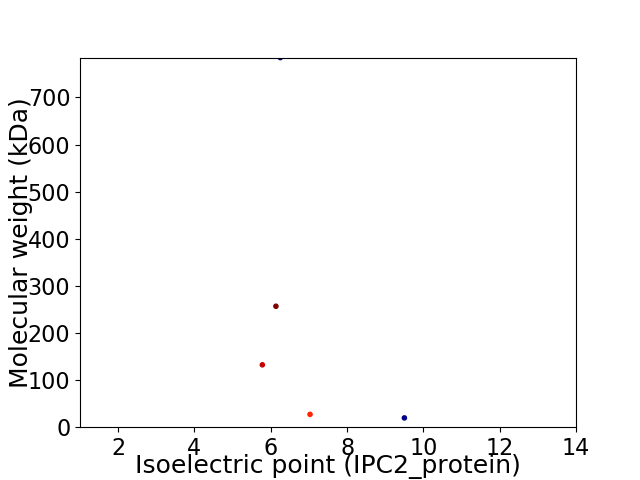
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E3J9S7|A0A0E3J9S7_9NIDO Putative nucleocapsid protein OS=Chinook salmon bafinivirus OX=1611837 PE=4 SV=1
MM1 pKa = 7.9ANLLWLTLLITLGLVSAQINTTLPDD26 pKa = 4.03CPIPTCTYY34 pKa = 8.94TPPTTTPFDD43 pKa = 3.62HH44 pKa = 7.0TSLHH48 pKa = 6.6SFFTHH53 pKa = 5.74DD54 pKa = 2.97TNYY57 pKa = 11.12AFLLSLKK64 pKa = 10.54YY65 pKa = 10.64NGTFIPATMAMPANNLSQTYY85 pKa = 7.86TLNYY89 pKa = 9.68KK90 pKa = 10.18NVFVNVPDD98 pKa = 3.93SQLPPPQRR106 pKa = 11.84SNTQLAVVATTASCLTATYY125 pKa = 8.89TAIPGTIPALAATDD139 pKa = 3.9MYY141 pKa = 10.74CVKK144 pKa = 9.68FTQSSNFVYY153 pKa = 10.37LYY155 pKa = 8.22DD156 pKa = 3.75TTVNTPVGVLLKK168 pKa = 10.87NPTAPVVTTILSTIMVNRR186 pKa = 11.84PTTVSTSQLLSNIYY200 pKa = 10.41SLMRR204 pKa = 11.84TRR206 pKa = 11.84SNVIQQQFTCTSCLFAIDD224 pKa = 4.13ASTGRR229 pKa = 11.84AATSPTTHH237 pKa = 7.46LLNNWSPSFFEE248 pKa = 4.79TKK250 pKa = 9.38TAMLDD255 pKa = 3.5HH256 pKa = 7.3VSMNTIYY263 pKa = 9.17QTARR267 pKa = 11.84GVTRR271 pKa = 11.84EE272 pKa = 4.0TFTSYY277 pKa = 10.88ILNAAKK283 pKa = 9.01TAAYY287 pKa = 10.33ARR289 pKa = 11.84GSPAYY294 pKa = 10.28YY295 pKa = 9.51IFSFAAIARR304 pKa = 11.84YY305 pKa = 7.88HH306 pKa = 5.23QAMVTPYY313 pKa = 10.13NYY315 pKa = 10.49NIGKK319 pKa = 9.26LCEE322 pKa = 3.88ATAACCLSNGIEE334 pKa = 4.26HH335 pKa = 6.9FDD337 pKa = 3.03IYY339 pKa = 10.93TWLFHH344 pKa = 6.74YY345 pKa = 10.67YY346 pKa = 10.8GFMKK350 pKa = 10.43SKK352 pKa = 10.5VYY354 pKa = 10.99SEE356 pKa = 4.61TFTTNPVLTSVINLATSIKK375 pKa = 9.56GKK377 pKa = 10.02HH378 pKa = 5.83SSYY381 pKa = 11.27CDD383 pKa = 3.31LTPYY387 pKa = 10.96SSFTAYY393 pKa = 10.87NLIMLQDD400 pKa = 3.48VVEE403 pKa = 4.41QYY405 pKa = 10.92QGTRR409 pKa = 11.84TTSTYY414 pKa = 11.58ANNFLNLTDD423 pKa = 3.49INGYY427 pKa = 9.87LPYY430 pKa = 9.53STHH433 pKa = 6.42TPLPQNYY440 pKa = 7.98QNYY443 pKa = 7.91MPFATYY449 pKa = 11.08LLATFYY455 pKa = 11.36SSFTLQQKK463 pKa = 10.31FDD465 pKa = 3.45FHH467 pKa = 7.44RR468 pKa = 11.84FLQSVCYY475 pKa = 9.25YY476 pKa = 10.67DD477 pKa = 3.62SGFISRR483 pKa = 11.84NQAYY487 pKa = 10.13HH488 pKa = 6.38SICHH492 pKa = 6.88IITLKK497 pKa = 10.11TSSSVPYY504 pKa = 9.39PAEE507 pKa = 4.03LQWPFTALPSSGYY520 pKa = 10.59DD521 pKa = 3.17SYY523 pKa = 11.57PVSTGLGTVAVGNTNPLDD541 pKa = 3.83YY542 pKa = 10.86PGICYY547 pKa = 10.15QSVCLTHH554 pKa = 5.66PTINFRR560 pKa = 11.84LLLSEE565 pKa = 5.98LPIKK569 pKa = 10.36WEE571 pKa = 3.69VRR573 pKa = 11.84PEE575 pKa = 4.42GIYY578 pKa = 9.03VTSRR582 pKa = 11.84NDD584 pKa = 3.38PTCLNFKK591 pKa = 9.41TSQLGTVFITLYY603 pKa = 10.72DD604 pKa = 3.89SNAXCTLQQTTYY616 pKa = 10.73SRR618 pKa = 11.84SAVLRR623 pKa = 11.84QVNTFTPNATDD634 pKa = 3.54LVVPYY639 pKa = 10.41GKK641 pKa = 8.51TPIYY645 pKa = 10.52NGAYY649 pKa = 9.78NINSTASGAVSXSTNTHH666 pKa = 6.31YY667 pKa = 10.48IPRR670 pKa = 11.84LLTSVLTPQSDD681 pKa = 4.12YY682 pKa = 11.2KK683 pKa = 10.13ITPTTTNLEE692 pKa = 4.11PFVEE696 pKa = 4.47INGTTFYY703 pKa = 11.39YY704 pKa = 10.47DD705 pKa = 3.2ANQAYY710 pKa = 10.47VNTSDD715 pKa = 4.41VYY717 pKa = 10.42QANVDD722 pKa = 4.0FGLPQTFSLDD732 pKa = 3.52TSFLNCTAFLCASDD746 pKa = 4.14YY747 pKa = 11.45KK748 pKa = 10.89CIADD752 pKa = 4.3FSPICSATQPLIDD765 pKa = 5.38ALLQTYY771 pKa = 7.33TLYY774 pKa = 10.82NQQLTLYY781 pKa = 9.19NSSLLTIQAAANYY794 pKa = 10.06LYY796 pKa = 8.9DD797 pKa = 3.09TRR799 pKa = 11.84TRR801 pKa = 11.84TRR803 pKa = 11.84RR804 pKa = 11.84SLLSTVNLGYY814 pKa = 11.06GDD816 pKa = 4.95VFQQPAWVYY825 pKa = 10.45EE826 pKa = 4.14IGATAAIPMIGPAVSVGLLASRR848 pKa = 11.84VASIEE853 pKa = 3.93STLYY857 pKa = 10.83QITQGVKK864 pKa = 9.38EE865 pKa = 5.15SIMAMNTKK873 pKa = 9.88LDD875 pKa = 3.98KK876 pKa = 10.51IANILHH882 pKa = 6.39SDD884 pKa = 3.74IEE886 pKa = 4.39RR887 pKa = 11.84LGLTVNTLASSFNTFTKK904 pKa = 10.55QVTDD908 pKa = 3.39KK909 pKa = 10.64FASVDD914 pKa = 3.45VAFNDD919 pKa = 4.54LSTSLNTLTTTTTVGASYY937 pKa = 10.35AAQINFATTTLTNLEE952 pKa = 4.21VQISQTTDD960 pKa = 2.68HH961 pKa = 6.98ALSCIAALNEE971 pKa = 4.4HH972 pKa = 6.37VLSPHH977 pKa = 6.93CISVSQLSAYY987 pKa = 8.28TPNALSNIGEE997 pKa = 4.39KK998 pKa = 10.54LVLTHH1003 pKa = 6.64YY1004 pKa = 10.85INGSLITFYY1013 pKa = 11.11HH1014 pKa = 6.95LIASKK1019 pKa = 10.58GVYY1022 pKa = 9.56RR1023 pKa = 11.84PLPKK1027 pKa = 9.67MYY1029 pKa = 10.51NSTHH1033 pKa = 6.38HH1034 pKa = 6.71LVATTGFYY1042 pKa = 10.47NGHH1045 pKa = 6.93CIFCGEE1051 pKa = 4.56HH1052 pKa = 5.81YY1053 pKa = 10.62CDD1055 pKa = 3.65ISVSVPCDD1063 pKa = 3.23YY1064 pKa = 10.91AYY1066 pKa = 10.25EE1067 pKa = 4.2PLKK1070 pKa = 10.55STVLAAYY1077 pKa = 8.35PLTTGVALLTLDD1089 pKa = 3.47STATNFIINYY1099 pKa = 9.02NNAPLISSSVPQPPIINNTDD1119 pKa = 3.09GTVITLQQQIIQLQTQFDD1137 pKa = 4.07HH1138 pKa = 7.17FNISSNFHH1146 pKa = 5.78VSEE1149 pKa = 3.92IQPIIDD1155 pKa = 3.87YY1156 pKa = 11.06YY1157 pKa = 10.58NAQINSSLNQIVDD1170 pKa = 4.26FGNGSTSNVGTIILIVLVVALALAAAIAIICGLKK1204 pKa = 10.12HH1205 pKa = 5.93
MM1 pKa = 7.9ANLLWLTLLITLGLVSAQINTTLPDD26 pKa = 4.03CPIPTCTYY34 pKa = 8.94TPPTTTPFDD43 pKa = 3.62HH44 pKa = 7.0TSLHH48 pKa = 6.6SFFTHH53 pKa = 5.74DD54 pKa = 2.97TNYY57 pKa = 11.12AFLLSLKK64 pKa = 10.54YY65 pKa = 10.64NGTFIPATMAMPANNLSQTYY85 pKa = 7.86TLNYY89 pKa = 9.68KK90 pKa = 10.18NVFVNVPDD98 pKa = 3.93SQLPPPQRR106 pKa = 11.84SNTQLAVVATTASCLTATYY125 pKa = 8.89TAIPGTIPALAATDD139 pKa = 3.9MYY141 pKa = 10.74CVKK144 pKa = 9.68FTQSSNFVYY153 pKa = 10.37LYY155 pKa = 8.22DD156 pKa = 3.75TTVNTPVGVLLKK168 pKa = 10.87NPTAPVVTTILSTIMVNRR186 pKa = 11.84PTTVSTSQLLSNIYY200 pKa = 10.41SLMRR204 pKa = 11.84TRR206 pKa = 11.84SNVIQQQFTCTSCLFAIDD224 pKa = 4.13ASTGRR229 pKa = 11.84AATSPTTHH237 pKa = 7.46LLNNWSPSFFEE248 pKa = 4.79TKK250 pKa = 9.38TAMLDD255 pKa = 3.5HH256 pKa = 7.3VSMNTIYY263 pKa = 9.17QTARR267 pKa = 11.84GVTRR271 pKa = 11.84EE272 pKa = 4.0TFTSYY277 pKa = 10.88ILNAAKK283 pKa = 9.01TAAYY287 pKa = 10.33ARR289 pKa = 11.84GSPAYY294 pKa = 10.28YY295 pKa = 9.51IFSFAAIARR304 pKa = 11.84YY305 pKa = 7.88HH306 pKa = 5.23QAMVTPYY313 pKa = 10.13NYY315 pKa = 10.49NIGKK319 pKa = 9.26LCEE322 pKa = 3.88ATAACCLSNGIEE334 pKa = 4.26HH335 pKa = 6.9FDD337 pKa = 3.03IYY339 pKa = 10.93TWLFHH344 pKa = 6.74YY345 pKa = 10.67YY346 pKa = 10.8GFMKK350 pKa = 10.43SKK352 pKa = 10.5VYY354 pKa = 10.99SEE356 pKa = 4.61TFTTNPVLTSVINLATSIKK375 pKa = 9.56GKK377 pKa = 10.02HH378 pKa = 5.83SSYY381 pKa = 11.27CDD383 pKa = 3.31LTPYY387 pKa = 10.96SSFTAYY393 pKa = 10.87NLIMLQDD400 pKa = 3.48VVEE403 pKa = 4.41QYY405 pKa = 10.92QGTRR409 pKa = 11.84TTSTYY414 pKa = 11.58ANNFLNLTDD423 pKa = 3.49INGYY427 pKa = 9.87LPYY430 pKa = 9.53STHH433 pKa = 6.42TPLPQNYY440 pKa = 7.98QNYY443 pKa = 7.91MPFATYY449 pKa = 11.08LLATFYY455 pKa = 11.36SSFTLQQKK463 pKa = 10.31FDD465 pKa = 3.45FHH467 pKa = 7.44RR468 pKa = 11.84FLQSVCYY475 pKa = 9.25YY476 pKa = 10.67DD477 pKa = 3.62SGFISRR483 pKa = 11.84NQAYY487 pKa = 10.13HH488 pKa = 6.38SICHH492 pKa = 6.88IITLKK497 pKa = 10.11TSSSVPYY504 pKa = 9.39PAEE507 pKa = 4.03LQWPFTALPSSGYY520 pKa = 10.59DD521 pKa = 3.17SYY523 pKa = 11.57PVSTGLGTVAVGNTNPLDD541 pKa = 3.83YY542 pKa = 10.86PGICYY547 pKa = 10.15QSVCLTHH554 pKa = 5.66PTINFRR560 pKa = 11.84LLLSEE565 pKa = 5.98LPIKK569 pKa = 10.36WEE571 pKa = 3.69VRR573 pKa = 11.84PEE575 pKa = 4.42GIYY578 pKa = 9.03VTSRR582 pKa = 11.84NDD584 pKa = 3.38PTCLNFKK591 pKa = 9.41TSQLGTVFITLYY603 pKa = 10.72DD604 pKa = 3.89SNAXCTLQQTTYY616 pKa = 10.73SRR618 pKa = 11.84SAVLRR623 pKa = 11.84QVNTFTPNATDD634 pKa = 3.54LVVPYY639 pKa = 10.41GKK641 pKa = 8.51TPIYY645 pKa = 10.52NGAYY649 pKa = 9.78NINSTASGAVSXSTNTHH666 pKa = 6.31YY667 pKa = 10.48IPRR670 pKa = 11.84LLTSVLTPQSDD681 pKa = 4.12YY682 pKa = 11.2KK683 pKa = 10.13ITPTTTNLEE692 pKa = 4.11PFVEE696 pKa = 4.47INGTTFYY703 pKa = 11.39YY704 pKa = 10.47DD705 pKa = 3.2ANQAYY710 pKa = 10.47VNTSDD715 pKa = 4.41VYY717 pKa = 10.42QANVDD722 pKa = 4.0FGLPQTFSLDD732 pKa = 3.52TSFLNCTAFLCASDD746 pKa = 4.14YY747 pKa = 11.45KK748 pKa = 10.89CIADD752 pKa = 4.3FSPICSATQPLIDD765 pKa = 5.38ALLQTYY771 pKa = 7.33TLYY774 pKa = 10.82NQQLTLYY781 pKa = 9.19NSSLLTIQAAANYY794 pKa = 10.06LYY796 pKa = 8.9DD797 pKa = 3.09TRR799 pKa = 11.84TRR801 pKa = 11.84TRR803 pKa = 11.84RR804 pKa = 11.84SLLSTVNLGYY814 pKa = 11.06GDD816 pKa = 4.95VFQQPAWVYY825 pKa = 10.45EE826 pKa = 4.14IGATAAIPMIGPAVSVGLLASRR848 pKa = 11.84VASIEE853 pKa = 3.93STLYY857 pKa = 10.83QITQGVKK864 pKa = 9.38EE865 pKa = 5.15SIMAMNTKK873 pKa = 9.88LDD875 pKa = 3.98KK876 pKa = 10.51IANILHH882 pKa = 6.39SDD884 pKa = 3.74IEE886 pKa = 4.39RR887 pKa = 11.84LGLTVNTLASSFNTFTKK904 pKa = 10.55QVTDD908 pKa = 3.39KK909 pKa = 10.64FASVDD914 pKa = 3.45VAFNDD919 pKa = 4.54LSTSLNTLTTTTTVGASYY937 pKa = 10.35AAQINFATTTLTNLEE952 pKa = 4.21VQISQTTDD960 pKa = 2.68HH961 pKa = 6.98ALSCIAALNEE971 pKa = 4.4HH972 pKa = 6.37VLSPHH977 pKa = 6.93CISVSQLSAYY987 pKa = 8.28TPNALSNIGEE997 pKa = 4.39KK998 pKa = 10.54LVLTHH1003 pKa = 6.64YY1004 pKa = 10.85INGSLITFYY1013 pKa = 11.11HH1014 pKa = 6.95LIASKK1019 pKa = 10.58GVYY1022 pKa = 9.56RR1023 pKa = 11.84PLPKK1027 pKa = 9.67MYY1029 pKa = 10.51NSTHH1033 pKa = 6.38HH1034 pKa = 6.71LVATTGFYY1042 pKa = 10.47NGHH1045 pKa = 6.93CIFCGEE1051 pKa = 4.56HH1052 pKa = 5.81YY1053 pKa = 10.62CDD1055 pKa = 3.65ISVSVPCDD1063 pKa = 3.23YY1064 pKa = 10.91AYY1066 pKa = 10.25EE1067 pKa = 4.2PLKK1070 pKa = 10.55STVLAAYY1077 pKa = 8.35PLTTGVALLTLDD1089 pKa = 3.47STATNFIINYY1099 pKa = 9.02NNAPLISSSVPQPPIINNTDD1119 pKa = 3.09GTVITLQQQIIQLQTQFDD1137 pKa = 4.07HH1138 pKa = 7.17FNISSNFHH1146 pKa = 5.78VSEE1149 pKa = 3.92IQPIIDD1155 pKa = 3.87YY1156 pKa = 11.06YY1157 pKa = 10.58NAQINSSLNQIVDD1170 pKa = 4.26FGNGSTSNVGTIILIVLVVALALAAAIAIICGLKK1204 pKa = 10.12HH1205 pKa = 5.93
Molecular weight: 132.39 kDa
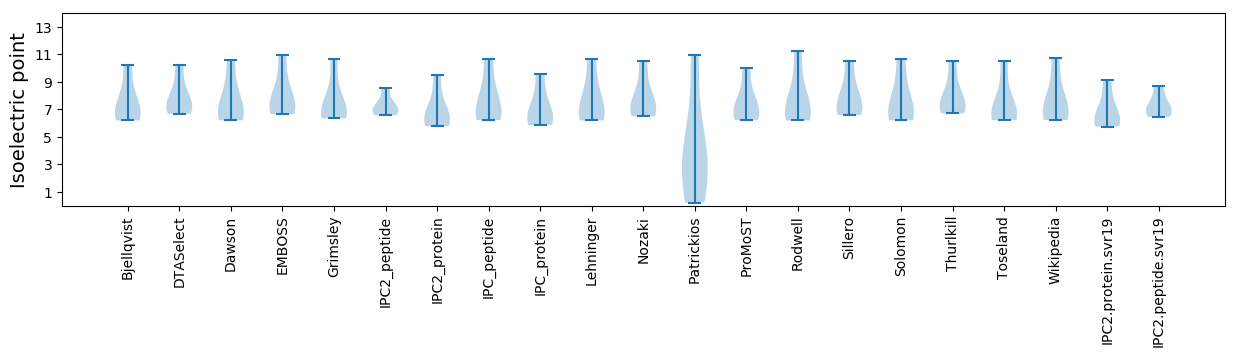
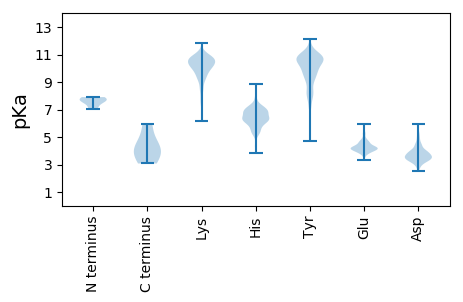
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3J9X8|A0A0E3J9X8_9NIDO Membrane protein OS=Chinook salmon bafinivirus OX=1611837 PE=4 SV=1
MM1 pKa = 7.56AFYY4 pKa = 9.94GPPMMPMSMPASFPPRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APKK25 pKa = 9.86KK26 pKa = 10.13RR27 pKa = 11.84PQQKK31 pKa = 9.51KK32 pKa = 7.16QPKK35 pKa = 8.17PQPKK39 pKa = 9.39PLNDD43 pKa = 3.56QQKK46 pKa = 7.53TAVAKK51 pKa = 9.94EE52 pKa = 4.32AIAKK56 pKa = 7.53ATTVPMPPMAPVVKK70 pKa = 9.83TSLRR74 pKa = 11.84TQVIKK79 pKa = 10.84GKK81 pKa = 10.44RR82 pKa = 11.84GEE84 pKa = 4.1LQDD87 pKa = 3.2ARR89 pKa = 11.84GRR91 pKa = 11.84LTQSEE96 pKa = 4.26INQLYY101 pKa = 9.01FQIATALNSGLGEE114 pKa = 4.04IGIHH118 pKa = 6.04NAATQEE124 pKa = 4.05GVRR127 pKa = 11.84LIVRR131 pKa = 11.84LNLAPPNSIAEE142 pKa = 4.29KK143 pKa = 10.02LYY145 pKa = 10.31KK146 pKa = 10.24SHH148 pKa = 7.25AAAHH152 pKa = 6.14LNSEE156 pKa = 4.18ALEE159 pKa = 4.43SKK161 pKa = 10.49FKK163 pKa = 10.71EE164 pKa = 4.26RR165 pKa = 11.84EE166 pKa = 3.9SDD168 pKa = 3.67PLFHH172 pKa = 7.78FSTTPEE178 pKa = 3.71
MM1 pKa = 7.56AFYY4 pKa = 9.94GPPMMPMSMPASFPPRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APKK25 pKa = 9.86KK26 pKa = 10.13RR27 pKa = 11.84PQQKK31 pKa = 9.51KK32 pKa = 7.16QPKK35 pKa = 8.17PQPKK39 pKa = 9.39PLNDD43 pKa = 3.56QQKK46 pKa = 7.53TAVAKK51 pKa = 9.94EE52 pKa = 4.32AIAKK56 pKa = 7.53ATTVPMPPMAPVVKK70 pKa = 9.83TSLRR74 pKa = 11.84TQVIKK79 pKa = 10.84GKK81 pKa = 10.44RR82 pKa = 11.84GEE84 pKa = 4.1LQDD87 pKa = 3.2ARR89 pKa = 11.84GRR91 pKa = 11.84LTQSEE96 pKa = 4.26INQLYY101 pKa = 9.01FQIATALNSGLGEE114 pKa = 4.04IGIHH118 pKa = 6.04NAATQEE124 pKa = 4.05GVRR127 pKa = 11.84LIVRR131 pKa = 11.84LNLAPPNSIAEE142 pKa = 4.29KK143 pKa = 10.02LYY145 pKa = 10.31KK146 pKa = 10.24SHH148 pKa = 7.25AAAHH152 pKa = 6.14LNSEE156 pKa = 4.18ALEE159 pKa = 4.43SKK161 pKa = 10.49FKK163 pKa = 10.71EE164 pKa = 4.26RR165 pKa = 11.84EE166 pKa = 3.9SDD168 pKa = 3.67PLFHH172 pKa = 7.78FSTTPEE178 pKa = 3.71
Molecular weight: 19.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10919 |
178 |
7011 |
2183.8 |
244.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.896 ± 0.283 | 1.951 ± 0.18 |
4.515 ± 0.361 | 3.627 ± 0.419 |
4.506 ± 0.169 | 4.277 ± 0.197 |
3.7 ± 0.377 | 6.768 ± 0.155 |
4.671 ± 0.524 | 9.387 ± 0.403 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.914 ± 0.225 | 5.696 ± 0.277 |
5.761 ± 0.254 | 4.185 ± 0.153 |
2.931 ± 0.217 | 7.217 ± 0.394 |
10.505 ± 0.702 | 5.843 ± 0.16 |
0.632 ± 0.035 | 4.991 ± 0.372 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
