
Prauserella marina
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Prauserella
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
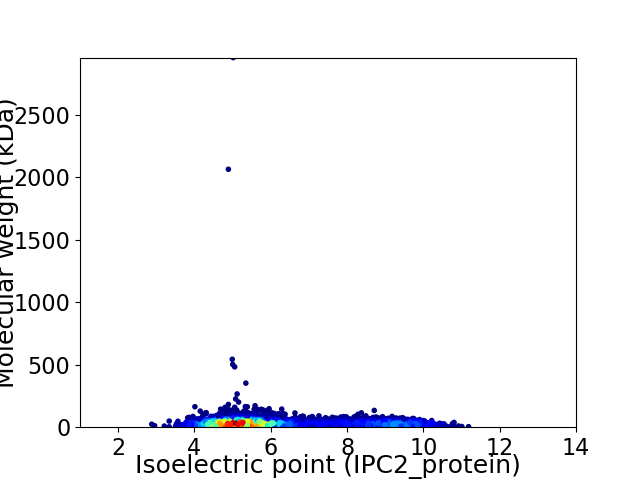
Virtual 2D-PAGE plot for 6551 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A222VRQ5|A0A222VRQ5_9PSEU Uncharacterized protein OS=Prauserella marina OX=530584 GN=DES30_108180 PE=4 SV=1
MM1 pKa = 7.88AIRR4 pKa = 11.84RR5 pKa = 11.84GSAALLSLLVALVWLVPSAFADD27 pKa = 3.78DD28 pKa = 4.74AKK30 pKa = 11.13DD31 pKa = 3.2AGRR34 pKa = 11.84PSVPQTQQVQRR45 pKa = 11.84AQVQPAQAQVLTWTGNNSLNEE66 pKa = 4.04YY67 pKa = 10.73GEE69 pKa = 4.74APTTATAGAATLVFEE84 pKa = 4.65NSVATGNTSGMTHH97 pKa = 6.95TLTFDD102 pKa = 3.29TSTPGYY108 pKa = 9.66NHH110 pKa = 7.2DD111 pKa = 3.63VDD113 pKa = 3.99VDD115 pKa = 3.85IVASPFDD122 pKa = 4.2ANGGRR127 pKa = 11.84HH128 pKa = 4.82EE129 pKa = 4.32VQVNLSPGKK138 pKa = 10.47YY139 pKa = 9.0RR140 pKa = 11.84FFCAIPGHH148 pKa = 5.17QQMQGVLTVTEE159 pKa = 4.69GGEE162 pKa = 4.12QDD164 pKa = 3.29TTAPTVTAEE173 pKa = 4.01VTGDD177 pKa = 3.08QDD179 pKa = 5.09AEE181 pKa = 4.29GNYY184 pKa = 10.09LGSATVSVSAEE195 pKa = 3.88DD196 pKa = 3.96AEE198 pKa = 4.66SGVAGIEE205 pKa = 4.24YY206 pKa = 10.47AVGEE210 pKa = 4.67GDD212 pKa = 3.63FQPYY216 pKa = 9.8SEE218 pKa = 4.82PVTLAEE224 pKa = 4.69PGDD227 pKa = 3.71YY228 pKa = 9.53TVQYY232 pKa = 10.36RR233 pKa = 11.84ATDD236 pKa = 3.59NAGNTSDD243 pKa = 4.03PGSVSFTVAEE253 pKa = 4.78GEE255 pKa = 4.55PEE257 pKa = 4.83DD258 pKa = 3.82NTPPEE263 pKa = 4.25VTAEE267 pKa = 3.95LAGNQDD273 pKa = 2.76ADD275 pKa = 3.83GNYY278 pKa = 10.2LGAATVILNAQDD290 pKa = 3.56AGSGVAGIEE299 pKa = 3.85YY300 pKa = 10.41DD301 pKa = 3.54LDD303 pKa = 3.87GAGFADD309 pKa = 3.81YY310 pKa = 10.87SEE312 pKa = 4.3PVRR315 pKa = 11.84VTEE318 pKa = 4.55LGEE321 pKa = 3.88HH322 pKa = 4.92TVAYY326 pKa = 9.7RR327 pKa = 11.84ATDD330 pKa = 3.21NAGNVSEE337 pKa = 5.07AGSATFTVIEE347 pKa = 4.75GSQDD351 pKa = 3.36DD352 pKa = 4.25TTPPHH357 pKa = 6.93VMAEE361 pKa = 4.11VTGQQDD367 pKa = 2.91GDD369 pKa = 3.86GNYY372 pKa = 10.15VGSAAVNLTAHH383 pKa = 6.83DD384 pKa = 4.39AEE386 pKa = 4.86SGVDD390 pKa = 3.46TVEE393 pKa = 3.93YY394 pKa = 10.73SLDD397 pKa = 3.11GGMYY401 pKa = 9.3TPYY404 pKa = 9.84TEE406 pKa = 4.25PVVVSAIGSHH416 pKa = 3.98TVEE419 pKa = 4.43YY420 pKa = 10.39RR421 pKa = 11.84ATDD424 pKa = 3.57LAGNTSEE431 pKa = 4.47PGSVSFAVVEE441 pKa = 4.17GQPDD445 pKa = 3.48DD446 pKa = 3.61TAPTVTAEE454 pKa = 4.15VTGEE458 pKa = 3.81QDD460 pKa = 3.14GDD462 pKa = 3.75GAYY465 pKa = 10.53LGAATVVISAEE476 pKa = 4.05DD477 pKa = 3.66AEE479 pKa = 4.77SGVASVEE486 pKa = 4.19YY487 pKa = 10.7ALGEE491 pKa = 4.45SEE493 pKa = 4.12FTGYY497 pKa = 9.13TDD499 pKa = 3.25PVEE502 pKa = 4.06VTEE505 pKa = 4.61PGEE508 pKa = 3.88HH509 pKa = 4.44TVAYY513 pKa = 9.65RR514 pKa = 11.84ATDD517 pKa = 3.21NAGNVSEE524 pKa = 4.75AGSVTFTVVEE534 pKa = 4.91GSQEE538 pKa = 3.91DD539 pKa = 3.92TTAPEE544 pKa = 3.57TSIRR548 pKa = 11.84ITGEE552 pKa = 3.78QDD554 pKa = 3.15WEE556 pKa = 3.72WSYY559 pKa = 11.97IDD561 pKa = 4.0LAKK564 pKa = 9.61ITLLAEE570 pKa = 4.66DD571 pKa = 5.08DD572 pKa = 4.08EE573 pKa = 5.64SGVALIEE580 pKa = 4.01YY581 pKa = 10.25SFGEE585 pKa = 4.56GSFRR589 pKa = 11.84PYY591 pKa = 10.46RR592 pKa = 11.84IPLTVLDD599 pKa = 4.32PGEE602 pKa = 4.26YY603 pKa = 7.63TVHH606 pKa = 6.42YY607 pKa = 9.63RR608 pKa = 11.84ATDD611 pKa = 3.41NAGNTSEE618 pKa = 4.46VGTARR623 pKa = 11.84FRR625 pKa = 11.84IVADD629 pKa = 4.21GVHH632 pKa = 5.81CTVSDD637 pKa = 3.35ARR639 pKa = 11.84EE640 pKa = 4.04TVVVGDD646 pKa = 3.93TDD648 pKa = 3.58TGVANVDD655 pKa = 3.53TGNGCTINDD664 pKa = 5.3LIVEE668 pKa = 4.25EE669 pKa = 5.02AGYY672 pKa = 9.45PDD674 pKa = 3.08QAAFVSHH681 pKa = 5.83VRR683 pKa = 11.84QVARR687 pKa = 11.84DD688 pKa = 3.77LEE690 pKa = 4.38AQGVITAQEE699 pKa = 3.76RR700 pKa = 11.84KK701 pKa = 10.02AIVSAARR708 pKa = 11.84GVPVPSAAA716 pKa = 3.86
MM1 pKa = 7.88AIRR4 pKa = 11.84RR5 pKa = 11.84GSAALLSLLVALVWLVPSAFADD27 pKa = 3.78DD28 pKa = 4.74AKK30 pKa = 11.13DD31 pKa = 3.2AGRR34 pKa = 11.84PSVPQTQQVQRR45 pKa = 11.84AQVQPAQAQVLTWTGNNSLNEE66 pKa = 4.04YY67 pKa = 10.73GEE69 pKa = 4.74APTTATAGAATLVFEE84 pKa = 4.65NSVATGNTSGMTHH97 pKa = 6.95TLTFDD102 pKa = 3.29TSTPGYY108 pKa = 9.66NHH110 pKa = 7.2DD111 pKa = 3.63VDD113 pKa = 3.99VDD115 pKa = 3.85IVASPFDD122 pKa = 4.2ANGGRR127 pKa = 11.84HH128 pKa = 4.82EE129 pKa = 4.32VQVNLSPGKK138 pKa = 10.47YY139 pKa = 9.0RR140 pKa = 11.84FFCAIPGHH148 pKa = 5.17QQMQGVLTVTEE159 pKa = 4.69GGEE162 pKa = 4.12QDD164 pKa = 3.29TTAPTVTAEE173 pKa = 4.01VTGDD177 pKa = 3.08QDD179 pKa = 5.09AEE181 pKa = 4.29GNYY184 pKa = 10.09LGSATVSVSAEE195 pKa = 3.88DD196 pKa = 3.96AEE198 pKa = 4.66SGVAGIEE205 pKa = 4.24YY206 pKa = 10.47AVGEE210 pKa = 4.67GDD212 pKa = 3.63FQPYY216 pKa = 9.8SEE218 pKa = 4.82PVTLAEE224 pKa = 4.69PGDD227 pKa = 3.71YY228 pKa = 9.53TVQYY232 pKa = 10.36RR233 pKa = 11.84ATDD236 pKa = 3.59NAGNTSDD243 pKa = 4.03PGSVSFTVAEE253 pKa = 4.78GEE255 pKa = 4.55PEE257 pKa = 4.83DD258 pKa = 3.82NTPPEE263 pKa = 4.25VTAEE267 pKa = 3.95LAGNQDD273 pKa = 2.76ADD275 pKa = 3.83GNYY278 pKa = 10.2LGAATVILNAQDD290 pKa = 3.56AGSGVAGIEE299 pKa = 3.85YY300 pKa = 10.41DD301 pKa = 3.54LDD303 pKa = 3.87GAGFADD309 pKa = 3.81YY310 pKa = 10.87SEE312 pKa = 4.3PVRR315 pKa = 11.84VTEE318 pKa = 4.55LGEE321 pKa = 3.88HH322 pKa = 4.92TVAYY326 pKa = 9.7RR327 pKa = 11.84ATDD330 pKa = 3.21NAGNVSEE337 pKa = 5.07AGSATFTVIEE347 pKa = 4.75GSQDD351 pKa = 3.36DD352 pKa = 4.25TTPPHH357 pKa = 6.93VMAEE361 pKa = 4.11VTGQQDD367 pKa = 2.91GDD369 pKa = 3.86GNYY372 pKa = 10.15VGSAAVNLTAHH383 pKa = 6.83DD384 pKa = 4.39AEE386 pKa = 4.86SGVDD390 pKa = 3.46TVEE393 pKa = 3.93YY394 pKa = 10.73SLDD397 pKa = 3.11GGMYY401 pKa = 9.3TPYY404 pKa = 9.84TEE406 pKa = 4.25PVVVSAIGSHH416 pKa = 3.98TVEE419 pKa = 4.43YY420 pKa = 10.39RR421 pKa = 11.84ATDD424 pKa = 3.57LAGNTSEE431 pKa = 4.47PGSVSFAVVEE441 pKa = 4.17GQPDD445 pKa = 3.48DD446 pKa = 3.61TAPTVTAEE454 pKa = 4.15VTGEE458 pKa = 3.81QDD460 pKa = 3.14GDD462 pKa = 3.75GAYY465 pKa = 10.53LGAATVVISAEE476 pKa = 4.05DD477 pKa = 3.66AEE479 pKa = 4.77SGVASVEE486 pKa = 4.19YY487 pKa = 10.7ALGEE491 pKa = 4.45SEE493 pKa = 4.12FTGYY497 pKa = 9.13TDD499 pKa = 3.25PVEE502 pKa = 4.06VTEE505 pKa = 4.61PGEE508 pKa = 3.88HH509 pKa = 4.44TVAYY513 pKa = 9.65RR514 pKa = 11.84ATDD517 pKa = 3.21NAGNVSEE524 pKa = 4.75AGSVTFTVVEE534 pKa = 4.91GSQEE538 pKa = 3.91DD539 pKa = 3.92TTAPEE544 pKa = 3.57TSIRR548 pKa = 11.84ITGEE552 pKa = 3.78QDD554 pKa = 3.15WEE556 pKa = 3.72WSYY559 pKa = 11.97IDD561 pKa = 4.0LAKK564 pKa = 9.61ITLLAEE570 pKa = 4.66DD571 pKa = 5.08DD572 pKa = 4.08EE573 pKa = 5.64SGVALIEE580 pKa = 4.01YY581 pKa = 10.25SFGEE585 pKa = 4.56GSFRR589 pKa = 11.84PYY591 pKa = 10.46RR592 pKa = 11.84IPLTVLDD599 pKa = 4.32PGEE602 pKa = 4.26YY603 pKa = 7.63TVHH606 pKa = 6.42YY607 pKa = 9.63RR608 pKa = 11.84ATDD611 pKa = 3.41NAGNTSEE618 pKa = 4.46VGTARR623 pKa = 11.84FRR625 pKa = 11.84IVADD629 pKa = 4.21GVHH632 pKa = 5.81CTVSDD637 pKa = 3.35ARR639 pKa = 11.84EE640 pKa = 4.04TVVVGDD646 pKa = 3.93TDD648 pKa = 3.58TGVANVDD655 pKa = 3.53TGNGCTINDD664 pKa = 5.3LIVEE668 pKa = 4.25EE669 pKa = 5.02AGYY672 pKa = 9.45PDD674 pKa = 3.08QAAFVSHH681 pKa = 5.83VRR683 pKa = 11.84QVARR687 pKa = 11.84DD688 pKa = 3.77LEE690 pKa = 4.38AQGVITAQEE699 pKa = 3.76RR700 pKa = 11.84KK701 pKa = 10.02AIVSAARR708 pKa = 11.84GVPVPSAAA716 pKa = 3.86
Molecular weight: 74.31 kDa
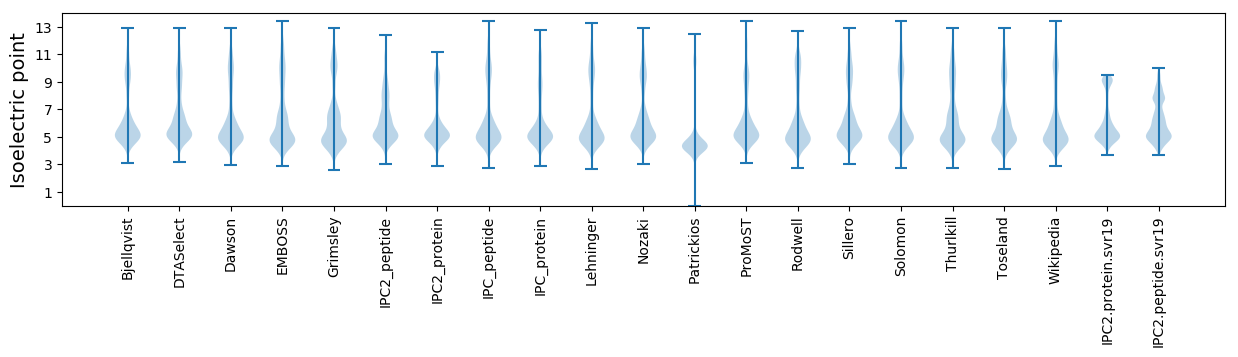
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G6P460|A0A1G6P460_9PSEU UPF0336 protein DES30_102934 OS=Prauserella marina OX=530584 GN=DES30_102934 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.72HH17 pKa = 5.63RR18 pKa = 11.84KK19 pKa = 7.56LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.71QGKK33 pKa = 8.65
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.72HH17 pKa = 5.63RR18 pKa = 11.84KK19 pKa = 7.56LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.71QGKK33 pKa = 8.65
Molecular weight: 4.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2104163 |
28 |
27313 |
321.2 |
34.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.102 ± 0.038 | 0.741 ± 0.013 |
5.914 ± 0.027 | 6.122 ± 0.028 |
2.904 ± 0.013 | 9.401 ± 0.028 |
2.148 ± 0.014 | 3.67 ± 0.021 |
2.138 ± 0.02 | 10.442 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.774 ± 0.013 | 1.942 ± 0.014 |
5.676 ± 0.026 | 2.618 ± 0.018 |
7.798 ± 0.034 | 5.362 ± 0.019 |
6.017 ± 0.021 | 8.833 ± 0.027 |
1.447 ± 0.012 | 1.951 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
