
Jingmen tombus-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.63
Get precalculated fractions of proteins
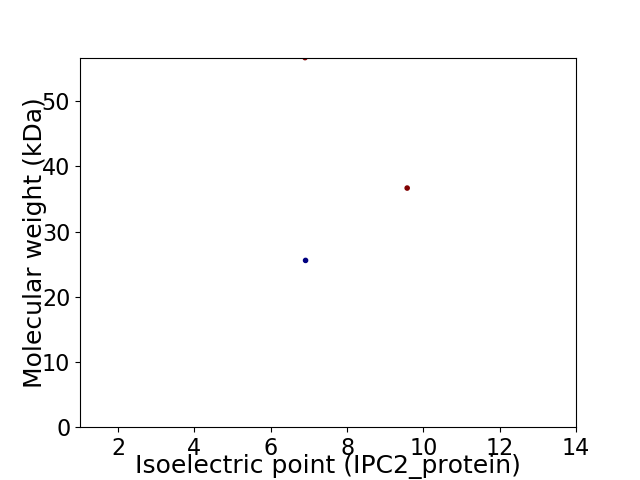
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
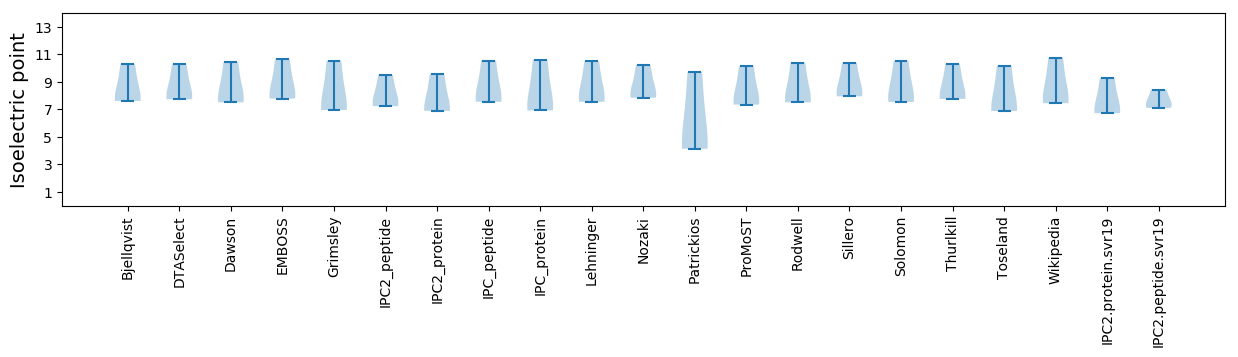
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG50|A0A1L3KG50_9VIRU RNA-directed RNA polymerase OS=Jingmen tombus-like virus 2 OX=1938656 PE=4 SV=1
MM1 pKa = 7.94RR2 pKa = 11.84DD3 pKa = 3.6YY4 pKa = 11.05EE5 pKa = 4.36ISNHH9 pKa = 5.96LNLPATRR16 pKa = 11.84PIKK19 pKa = 9.99WRR21 pKa = 11.84GQVLDD26 pKa = 5.15DD27 pKa = 4.55RR28 pKa = 11.84DD29 pKa = 3.74HH30 pKa = 6.69PAIIRR35 pKa = 11.84IGPSSGPQPYY45 pKa = 10.07APHH48 pKa = 6.84HH49 pKa = 6.02CWKK52 pKa = 10.09NWVRR56 pKa = 11.84GVKK59 pKa = 9.56KK60 pKa = 9.71RR61 pKa = 11.84CAHH64 pKa = 6.77AFHH67 pKa = 6.9PHH69 pKa = 6.97DD70 pKa = 4.19DD71 pKa = 3.68FEE73 pKa = 4.88RR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84FFTFVQSQIKK86 pKa = 8.57TMPRR90 pKa = 11.84LRR92 pKa = 11.84SGLKK96 pKa = 10.18YY97 pKa = 10.64EE98 pKa = 4.6SLLDD102 pKa = 3.45DD103 pKa = 4.24WLAHH107 pKa = 5.89SNYY110 pKa = 8.5TAARR114 pKa = 11.84AAEE117 pKa = 4.14LKK119 pKa = 10.58KK120 pKa = 10.71LASDD124 pKa = 3.94YY125 pKa = 10.9MNGDD129 pKa = 2.59IDD131 pKa = 3.62MNRR134 pKa = 11.84IYY136 pKa = 10.96LCDD139 pKa = 3.62SFIKK143 pKa = 10.55SEE145 pKa = 4.15MYY147 pKa = 10.74DD148 pKa = 3.52EE149 pKa = 4.49IKK151 pKa = 9.93EE152 pKa = 3.93ARR154 pKa = 11.84IINARR159 pKa = 11.84TDD161 pKa = 3.13WFKK164 pKa = 11.54VIAGPYY170 pKa = 7.51IHH172 pKa = 7.3AIEE175 pKa = 4.21KK176 pKa = 10.23LVYY179 pKa = 9.73DD180 pKa = 3.86DD181 pKa = 5.66HH182 pKa = 6.61FVKK185 pKa = 10.62HH186 pKa = 5.45KK187 pKa = 9.64TPAQIVKK194 pKa = 10.12RR195 pKa = 11.84LNKK198 pKa = 9.51LRR200 pKa = 11.84SVGSIFMEE208 pKa = 3.91TDD210 pKa = 3.05YY211 pKa = 11.78SSFEE215 pKa = 3.98GSFDD219 pKa = 3.93PEE221 pKa = 4.15LQRR224 pKa = 11.84HH225 pKa = 5.05VEE227 pKa = 3.9RR228 pKa = 11.84ALWDD232 pKa = 3.58HH233 pKa = 6.28MLVDD237 pKa = 4.08YY238 pKa = 10.82PEE240 pKa = 4.37ILSIIQRR247 pKa = 11.84AYY249 pKa = 9.16SRR251 pKa = 11.84SHH253 pKa = 5.68IRR255 pKa = 11.84YY256 pKa = 8.91KK257 pKa = 10.88NRR259 pKa = 11.84AHH261 pKa = 7.39AIFEE265 pKa = 4.2GSRR268 pKa = 11.84MSGDD272 pKa = 2.85MWTSLANGFTNKK284 pKa = 9.43MINEE288 pKa = 4.0YY289 pKa = 9.83MAMCARR295 pKa = 11.84DD296 pKa = 3.48NEE298 pKa = 3.93YY299 pKa = 10.95DD300 pKa = 3.88YY301 pKa = 11.61LVEE304 pKa = 6.1GDD306 pKa = 4.37DD307 pKa = 6.03AIICSSSRR315 pKa = 11.84WDD317 pKa = 3.24TDD319 pKa = 2.75IPARR323 pKa = 11.84LGFKK327 pKa = 9.5LTLEE331 pKa = 4.24TASDD335 pKa = 3.9FNEE338 pKa = 4.61LSFCGLNAVDD348 pKa = 5.51GVLIPNVRR356 pKa = 11.84KK357 pKa = 9.49ILTRR361 pKa = 11.84YY362 pKa = 10.21GYY364 pKa = 7.19TAEE367 pKa = 3.99MKK369 pKa = 10.69YY370 pKa = 11.31YY371 pKa = 10.15MDD373 pKa = 3.78VKK375 pKa = 10.26TNNQRR380 pKa = 11.84YY381 pKa = 8.8RR382 pKa = 11.84DD383 pKa = 3.78LMYY386 pKa = 11.08SKK388 pKa = 10.83ACSLLATSAKK398 pKa = 10.14VPVLGEE404 pKa = 3.94LARR407 pKa = 11.84QQMRR411 pKa = 11.84VWNGVNNFRR420 pKa = 11.84YY421 pKa = 8.83YY422 pKa = 10.98DD423 pKa = 2.72WWEE426 pKa = 3.89RR427 pKa = 11.84QFYY430 pKa = 10.47SYY432 pKa = 11.25DD433 pKa = 3.53EE434 pKa = 4.57PEE436 pKa = 4.56PLQSQPISMSVRR448 pKa = 11.84RR449 pKa = 11.84FVEE452 pKa = 4.31EE453 pKa = 4.78KK454 pKa = 8.54YY455 pKa = 10.56HH456 pKa = 7.89IPIVLQLQLEE466 pKa = 4.33EE467 pKa = 5.28EE468 pKa = 4.2IRR470 pKa = 11.84CCNFKK475 pKa = 11.05VFDD478 pKa = 3.84IEE480 pKa = 4.21FF481 pKa = 4.1
MM1 pKa = 7.94RR2 pKa = 11.84DD3 pKa = 3.6YY4 pKa = 11.05EE5 pKa = 4.36ISNHH9 pKa = 5.96LNLPATRR16 pKa = 11.84PIKK19 pKa = 9.99WRR21 pKa = 11.84GQVLDD26 pKa = 5.15DD27 pKa = 4.55RR28 pKa = 11.84DD29 pKa = 3.74HH30 pKa = 6.69PAIIRR35 pKa = 11.84IGPSSGPQPYY45 pKa = 10.07APHH48 pKa = 6.84HH49 pKa = 6.02CWKK52 pKa = 10.09NWVRR56 pKa = 11.84GVKK59 pKa = 9.56KK60 pKa = 9.71RR61 pKa = 11.84CAHH64 pKa = 6.77AFHH67 pKa = 6.9PHH69 pKa = 6.97DD70 pKa = 4.19DD71 pKa = 3.68FEE73 pKa = 4.88RR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84FFTFVQSQIKK86 pKa = 8.57TMPRR90 pKa = 11.84LRR92 pKa = 11.84SGLKK96 pKa = 10.18YY97 pKa = 10.64EE98 pKa = 4.6SLLDD102 pKa = 3.45DD103 pKa = 4.24WLAHH107 pKa = 5.89SNYY110 pKa = 8.5TAARR114 pKa = 11.84AAEE117 pKa = 4.14LKK119 pKa = 10.58KK120 pKa = 10.71LASDD124 pKa = 3.94YY125 pKa = 10.9MNGDD129 pKa = 2.59IDD131 pKa = 3.62MNRR134 pKa = 11.84IYY136 pKa = 10.96LCDD139 pKa = 3.62SFIKK143 pKa = 10.55SEE145 pKa = 4.15MYY147 pKa = 10.74DD148 pKa = 3.52EE149 pKa = 4.49IKK151 pKa = 9.93EE152 pKa = 3.93ARR154 pKa = 11.84IINARR159 pKa = 11.84TDD161 pKa = 3.13WFKK164 pKa = 11.54VIAGPYY170 pKa = 7.51IHH172 pKa = 7.3AIEE175 pKa = 4.21KK176 pKa = 10.23LVYY179 pKa = 9.73DD180 pKa = 3.86DD181 pKa = 5.66HH182 pKa = 6.61FVKK185 pKa = 10.62HH186 pKa = 5.45KK187 pKa = 9.64TPAQIVKK194 pKa = 10.12RR195 pKa = 11.84LNKK198 pKa = 9.51LRR200 pKa = 11.84SVGSIFMEE208 pKa = 3.91TDD210 pKa = 3.05YY211 pKa = 11.78SSFEE215 pKa = 3.98GSFDD219 pKa = 3.93PEE221 pKa = 4.15LQRR224 pKa = 11.84HH225 pKa = 5.05VEE227 pKa = 3.9RR228 pKa = 11.84ALWDD232 pKa = 3.58HH233 pKa = 6.28MLVDD237 pKa = 4.08YY238 pKa = 10.82PEE240 pKa = 4.37ILSIIQRR247 pKa = 11.84AYY249 pKa = 9.16SRR251 pKa = 11.84SHH253 pKa = 5.68IRR255 pKa = 11.84YY256 pKa = 8.91KK257 pKa = 10.88NRR259 pKa = 11.84AHH261 pKa = 7.39AIFEE265 pKa = 4.2GSRR268 pKa = 11.84MSGDD272 pKa = 2.85MWTSLANGFTNKK284 pKa = 9.43MINEE288 pKa = 4.0YY289 pKa = 9.83MAMCARR295 pKa = 11.84DD296 pKa = 3.48NEE298 pKa = 3.93YY299 pKa = 10.95DD300 pKa = 3.88YY301 pKa = 11.61LVEE304 pKa = 6.1GDD306 pKa = 4.37DD307 pKa = 6.03AIICSSSRR315 pKa = 11.84WDD317 pKa = 3.24TDD319 pKa = 2.75IPARR323 pKa = 11.84LGFKK327 pKa = 9.5LTLEE331 pKa = 4.24TASDD335 pKa = 3.9FNEE338 pKa = 4.61LSFCGLNAVDD348 pKa = 5.51GVLIPNVRR356 pKa = 11.84KK357 pKa = 9.49ILTRR361 pKa = 11.84YY362 pKa = 10.21GYY364 pKa = 7.19TAEE367 pKa = 3.99MKK369 pKa = 10.69YY370 pKa = 11.31YY371 pKa = 10.15MDD373 pKa = 3.78VKK375 pKa = 10.26TNNQRR380 pKa = 11.84YY381 pKa = 8.8RR382 pKa = 11.84DD383 pKa = 3.78LMYY386 pKa = 11.08SKK388 pKa = 10.83ACSLLATSAKK398 pKa = 10.14VPVLGEE404 pKa = 3.94LARR407 pKa = 11.84QQMRR411 pKa = 11.84VWNGVNNFRR420 pKa = 11.84YY421 pKa = 8.83YY422 pKa = 10.98DD423 pKa = 2.72WWEE426 pKa = 3.89RR427 pKa = 11.84QFYY430 pKa = 10.47SYY432 pKa = 11.25DD433 pKa = 3.53EE434 pKa = 4.57PEE436 pKa = 4.56PLQSQPISMSVRR448 pKa = 11.84RR449 pKa = 11.84FVEE452 pKa = 4.31EE453 pKa = 4.78KK454 pKa = 8.54YY455 pKa = 10.56HH456 pKa = 7.89IPIVLQLQLEE466 pKa = 4.33EE467 pKa = 5.28EE468 pKa = 4.2IRR470 pKa = 11.84CCNFKK475 pKa = 11.05VFDD478 pKa = 3.84IEE480 pKa = 4.21FF481 pKa = 4.1
Molecular weight: 56.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG11|A0A1L3KG11_9VIRU Uncharacterized protein OS=Jingmen tombus-like virus 2 OX=1938656 PE=4 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84KK3 pKa = 9.97ANLMNGASGPTIQSNNNNPPKK24 pKa = 10.76SNNDD28 pKa = 3.12ANRR31 pKa = 11.84VVPASRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84NRR43 pKa = 11.84NRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84APFTRR53 pKa = 11.84PSPPLAEE60 pKa = 4.12SASLNTYY67 pKa = 10.71ADD69 pKa = 3.16IVQRR73 pKa = 11.84GGMAILRR80 pKa = 11.84CRR82 pKa = 11.84EE83 pKa = 3.95VFQILAPLSEE93 pKa = 4.46SPIALMLPATPTKK106 pKa = 8.24WTRR109 pKa = 11.84TRR111 pKa = 11.84TGVLSSTYY119 pKa = 10.24AAYY122 pKa = 10.25RR123 pKa = 11.84PLQMRR128 pKa = 11.84LTYY131 pKa = 9.95VPSVAATTAGSVVVGTVFAGVRR153 pKa = 11.84LEE155 pKa = 4.15TTDD158 pKa = 3.77FNGLIRR164 pKa = 11.84QMPVSNGGFVTTVWHH179 pKa = 6.26SCTSRR184 pKa = 11.84VSLGTNLRR192 pKa = 11.84ANNFPTFSVDD202 pKa = 2.93MDD204 pKa = 5.97DD205 pKa = 3.38IPFWILVGTSSTVQPGWLVVEE226 pKa = 4.72AQFSLHH232 pKa = 6.56NPLNNQGAVSFSGFGPATFTQGTGNTTISIPQADD266 pKa = 3.8FVGDD270 pKa = 3.91LEE272 pKa = 4.45SGRR275 pKa = 11.84EE276 pKa = 4.02YY277 pKa = 11.28VFTPSASVGPSEE289 pKa = 4.53GVVAGTLEE297 pKa = 4.33PFRR300 pKa = 11.84AAYY303 pKa = 7.7TALTGDD309 pKa = 3.82NYY311 pKa = 10.84QFIGEE316 pKa = 4.05FSIPSTVTQFIAGVGRR332 pKa = 11.84SANFTQVEE340 pKa = 4.21
MM1 pKa = 7.86RR2 pKa = 11.84KK3 pKa = 9.97ANLMNGASGPTIQSNNNNPPKK24 pKa = 10.76SNNDD28 pKa = 3.12ANRR31 pKa = 11.84VVPASRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84NRR43 pKa = 11.84NRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84APFTRR53 pKa = 11.84PSPPLAEE60 pKa = 4.12SASLNTYY67 pKa = 10.71ADD69 pKa = 3.16IVQRR73 pKa = 11.84GGMAILRR80 pKa = 11.84CRR82 pKa = 11.84EE83 pKa = 3.95VFQILAPLSEE93 pKa = 4.46SPIALMLPATPTKK106 pKa = 8.24WTRR109 pKa = 11.84TRR111 pKa = 11.84TGVLSSTYY119 pKa = 10.24AAYY122 pKa = 10.25RR123 pKa = 11.84PLQMRR128 pKa = 11.84LTYY131 pKa = 9.95VPSVAATTAGSVVVGTVFAGVRR153 pKa = 11.84LEE155 pKa = 4.15TTDD158 pKa = 3.77FNGLIRR164 pKa = 11.84QMPVSNGGFVTTVWHH179 pKa = 6.26SCTSRR184 pKa = 11.84VSLGTNLRR192 pKa = 11.84ANNFPTFSVDD202 pKa = 2.93MDD204 pKa = 5.97DD205 pKa = 3.38IPFWILVGTSSTVQPGWLVVEE226 pKa = 4.72AQFSLHH232 pKa = 6.56NPLNNQGAVSFSGFGPATFTQGTGNTTISIPQADD266 pKa = 3.8FVGDD270 pKa = 3.91LEE272 pKa = 4.45SGRR275 pKa = 11.84EE276 pKa = 4.02YY277 pKa = 11.28VFTPSASVGPSEE289 pKa = 4.53GVVAGTLEE297 pKa = 4.33PFRR300 pKa = 11.84AAYY303 pKa = 7.7TALTGDD309 pKa = 3.82NYY311 pKa = 10.84QFIGEE316 pKa = 4.05FSIPSTVTQFIAGVGRR332 pKa = 11.84SANFTQVEE340 pKa = 4.21
Molecular weight: 36.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1051 |
230 |
481 |
350.3 |
39.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.517 ± 0.496 | 1.427 ± 0.302 |
5.233 ± 1.015 | 4.757 ± 0.758 |
4.948 ± 0.234 | 5.614 ± 0.981 |
2.188 ± 0.629 | 5.709 ± 0.697 |
3.33 ± 1.081 | 7.612 ± 0.637 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.854 ± 0.353 | 5.138 ± 0.607 |
5.519 ± 0.738 | 3.52 ± 0.211 |
7.802 ± 0.271 | 8.088 ± 0.635 |
7.041 ± 1.937 | 6.185 ± 0.95 |
1.522 ± 0.443 | 3.996 ± 0.845 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
